5F25
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1H
 
 | | Crystal structure of the BRD9 bromodamian in complex with BI-9564. | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1L
 
 | | Crystal structure of the bromodomain of BRD9 in complex with compound 9. | | Descriptor: | 5-[3,5-dimethoxy-4-[(3-oxidanylazetidin-1-yl)methyl]phenyl]-1,3-dimethyl-pyridin-2-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
4N4F
 
 | |
4N3W
 
 | |
5XHE
 
 | | Crystal structure analysis of the second bromodomain of BRD2 covalently linked to b-mercaptoethanol | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S.K, Deshmukh, P. | | Deposit date: | 2017-04-20 | | Release date: | 2017-09-06 | | Last modified: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5XHK
 
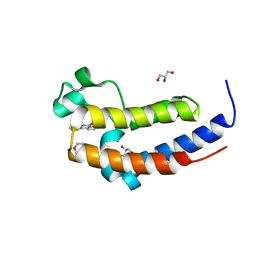 | | Crystal structure of the BRD2-BD2 in complex with phenanthridinone | | Descriptor: | Bromodomain-containing protein 2, GLYCEROL, METHOXYETHANE, ... | | Authors: | Padmanabhan, B, Mathur, S, Tripathi, S, Deshmukh, P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Insights into the crystal structure of BRD2-BD2 - phenanthridinone complex and theoretical studies on phenanthridinone analogs.
J. Biomol. Struct. Dyn., 36, 2018
|
|
5YE4
 
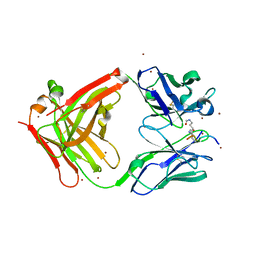 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | Descriptor: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
6ALC
 
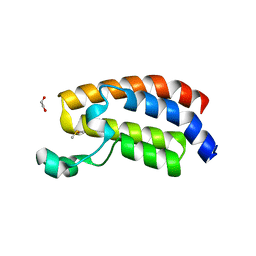 | | CREBBP bromodomain in complex with Cpd 4 (1-(1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[1-(cyclopropylmethyl)-3-(1H-indol-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]ethan-1-one, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | Design and synthesis of a biaryl series as inhibitors for the bromodomains of CBP/P300.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6AY5
 
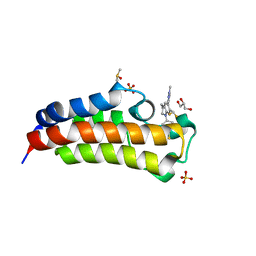 | | CREBBP bromodomain in complex with Cpd17 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-3-methylbenzo[d]thiazol-2(3H)-one) | | Descriptor: | 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-3-methyl-1,3-benzothiazol-2(3H)-one, CREB-binding protein, DIMETHYL SULFOXIDE, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
6AXQ
 
 | |
5YE3
 
 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
6AY3
 
 | | CREBBP bromodomain in complex with Cpd16 (5-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1H-indole-3-carboxamide) | | Descriptor: | 1,2-ETHANEDIOL, 5-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1H-indole-3-carboxamide, CREB-binding protein, ... | | Authors: | Murray, J.M. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.391 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
5DW1
 
 | | X-ray crystal structure of human BRD2(BD2) in complex with RVX297 to 1.55 A resolution | | Descriptor: | 2-{3,5-dimethyl-4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}-5,7-dimethoxyquinazolin-4(3H)-one, Bromodomain-containing protein 2, SODIUM ION | | Authors: | White, A, Fontano, E, Suto, R.K. | | Deposit date: | 2015-09-22 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | RVX-297- a novel BD2 selective inhibitor of BET bromodomains.
Biochem.Biophys.Res.Commun., 477, 2016
|
|
5EK9
 
 | | Crystal structure of the second bromodomain of human BRD2 in complex with a tetrahydroquinoline inhibitor | | Descriptor: | Bromodomain-containing protein 2, propan-2-yl ~{N}-[(2~{S},4~{R})-1-ethanoyl-6-(furan-2-yl)-2-methyl-3,4-dihydro-2~{H}-quinolin-4-yl]carbamate | | Authors: | Tallant, C, Slavish, P.J, Siejka, P, Bharatham, N, Shadrick, W.R, Chai, S, Young, B.M, Boyd, V.A, Heroven, C, Picaud, S, Fedorov, O, Chen, T, Lee, R.E, Guy, R.K, Shelat, A.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-03 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Exploiting a water network to achieve enthalpy-driven, bromodomain-selective BET inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
5W0E
 
 | | CREBBP bromodomain in complex with Cpd19 (3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-N-methyl-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide) | | Descriptor: | 3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-N-methyl-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A Unique Approach to Design Potent and Selective Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP) Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UEW
 
 | | BRD2 Bromodomain2 with A-1360579 | | Descriptor: | Bromodomain-containing protein 2, N-[3-(4-methoxy-1-methyl-6-oxo-1,6-dihydropyridin-3-yl)-4-phenoxyphenyl]methanesulfonamide | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
5TCH
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form, TrpA-G66V mutant | | Descriptor: | FORMIC ACID, MALONATE ION, Tryptophan synthase alpha chain, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5IX0
 
 | | HDAC2 WITH LIGAND BRD7232 | | Descriptor: | (3-exo)-N-(4-amino-4'-fluoro[1,1'-biphenyl]-3-yl)-8-oxabicyclo[3.2.1]octane-3-carboxamide, 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ... | | Authors: | Steinbacher, S. | | Deposit date: | 2016-03-23 | | Release date: | 2016-08-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Kinetic and structural insights into the binding of histone deacetylase 1 and 2 (HDAC1, 2) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5TCF
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form | | Descriptor: | FORMIC ACID, MALONATE ION, POTASSIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCG
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate-bound form | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, FORMIC ACID, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
