2VFD
 
 | | Crystal structure of the F96S mutant of Plasmodium falciparum triosephosphate isomerase | | Descriptor: | SULFATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Gayathri, P, Banerjee, M, Vijayalakshmi, A, Balaram, H, Balaram, P, Murthy, M.R.N. | | Deposit date: | 2007-11-03 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Biochemical and Structural Characterization of Residue 96 Mutants of Plasmodium Falciparum Triosephosphate Isomerase: Active-Site Loop Conformation, Hydration and Identification of a Dimer-Interface Ligand-Binding Site.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2HII
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
2HIK
 
 | | heterotrimeric PCNA sliding clamp | | Descriptor: | PCNA1 (SSO0397), PCNA2 (SSO1047), PCNA3 (SSO0405) | | Authors: | Pascal, J.M, Tsodikov, O.V, Ellenberger, T. | | Deposit date: | 2006-06-29 | | Release date: | 2006-11-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Flexible Interface between DNA Ligase and PCNA Supports Conformational Switching and Efficient Ligation of DNA.
Mol.Cell, 24, 2006
|
|
6U3K
 
 | | The crystal structure of 4-(pyridin-2-yl)benzoate-bound CYP199A4 | | Descriptor: | 4-(pyridin-2-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2019-08-21 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biophysical Techniques for Distinguishing Ligand Binding Modes in Cytochrome P450 Monooxygenases.
Biochemistry, 59, 2020
|
|
3K6P
 
 | | Estrogen Related Receptor alpha in Complex with an Ether Based Ligand | | Descriptor: | 4-(4-{[(5R)-2,4-dioxo-1,3-thiazolidin-5-yl]methyl}-2-methoxyphenoxy)-3-(trifluoromethyl)benzonitrile, Steroid hormone receptor ERR1 | | Authors: | Abad, M.C, Patch, R.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Development of Diaryl Ether based ligands for Estrogen Related Receptor alpha as Potential Anti-Diabetic Agents.
To be Published
|
|
1UEQ
 
 | | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein) | | Descriptor: | MEMBRANE ASSOCIATED GUANYLATE KINASE INVERTED-2 (MAGI-2) | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The First PDZ domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 protein)
To be Published
|
|
1UFW
 
 | | Solution structure of RNP domain in Synaptojanin 2 | | Descriptor: | Synaptojanin 2 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNP domain in Synaptojanin 2
To be Published
|
|
1UF1
 
 | | Solution structure of the second PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Saito, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second PDZ domain of human KIAA1526 protein
To be Published
|
|
4Z90
 
 | | ELIC bound with the anesthetic isoflurane in the resting state | | Descriptor: | (2R)-2-chloro-2-(difluoromethoxy)-1,1,1-trifluoroethane, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Chen, Q, Kinde, M, Arjunan, P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Direct Pore Binding as a Mechanism for Isoflurane Inhibition of the Pentameric Ligand-gated Ion Channel ELIC.
Sci Rep, 5, 2015
|
|
4WVW
 
 | | Chicken Galectin-8 N-terminal domain complexed with 3'-sialyl-lactose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Galectin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ruiz, F.M, Romero, A. | | Deposit date: | 2014-11-07 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combining Crystallography and Hydrogen-Deuterium Exchange to Study Galectin-Ligand Complexes.
Chemistry, 21, 2015
|
|
4U6D
 
 | | Zg3615, a family 117 glycoside hydrolase in complex with beta-3,6-anhydro-L-galactose | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-beta-L-galactopyranose, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6UD6
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | CHLORIDE ION, GLYCEROL, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
6UDC
 
 | | Spectroscopic and structural characterization of a genetically encoded direct sensor for protein-ligand interactions | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Mills, J.H, Gleason, P.R, Simmons, C.R, Henderson, J.N, Kartchner, B.K. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Origins of Altered Spectroscopic Properties upon Ligand Binding in Proteins Containing a Fluorescent Noncanonical Amino Acid.
Biochemistry, 60, 2021
|
|
1UFG
 
 | | Solution structure of immunoglobulin like domain of mouse nuclear lamin | | Descriptor: | Lamin A | | Authors: | Kobayashi, N, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-29 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of immunoglobulin like domain of mouse nuclear lamin
To be Published
|
|
1UDM
 
 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1UFN
 
 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
4U6B
 
 | | Zg3597, a family 117 glycoside hydrolase, produced by the marine bacterium Zobellia galactanivorans | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Ficko-Blean, E. | | Deposit date: | 2014-07-28 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Biochemical and structural investigation of two paralogous glycoside hydrolases from Zobellia galactanivorans: novel insights into the evolution, dimerization plasticity and catalytic mechanism of the GH117 family.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4U79
 
 | | Crystal structure of human JNK3 in complex with a benzenesulfonamide inhibitor. | | Descriptor: | Mitogen-activated protein kinase 10, N-{4-[(3-{2-[(trans-4-aminocyclohexyl)amino]pyrimidin-4-yl}pyridin-2-yl)oxy]naphthalen-1-yl}benzenesulfonamide | | Authors: | Mohr, C. | | Deposit date: | 2014-07-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Unfolded Protein Response in Cancer: IRE1 alpha Inhibition by Selective Kinase Ligands Does Not Impair Tumor Cell Viability.
Acs Med.Chem.Lett., 6, 2015
|
|
3SK7
 
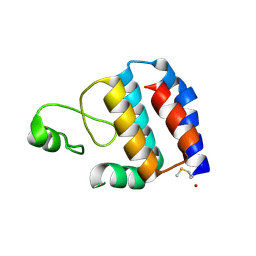 | | Crystal Structure of V. cholerae SeqA | | Descriptor: | Protein SeqA | | Authors: | Chung, Y.S, Guarne, A. | | Deposit date: | 2011-06-22 | | Release date: | 2012-12-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Investigating the aggregational property of SeqAvibrio in the absence of the oligomerization domain
To be Published
|
|
1UEP
 
 | | Solution Structure of The Third PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) | | Descriptor: | Membrane Associated Guanylate Kinase Inverted-2 (MAGI-2) | | Authors: | Miyamoto, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The Third PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein)
To be Published
|
|
3NY2
 
 | | Structure of the ubr-box of UBR2 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase UBR2, ZINC ION | | Authors: | Matta-Camacho, E, Kozlov, G, Li, F, Gehring, K. | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of substrate recognition and specificity in the N-end rule pathway.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1UF2
 
 | | The Atomic Structure of Rice dwarf Virus (RDV) | | Descriptor: | Core protein P3, Outer capsid protein P8, Structural protein P7 | | Authors: | Nakagawa, A, Miyazaki, N, Taka, J, Naitow, H, Ogawa, A, Fujimoto, Z, Mizuno, H, Higashi, T, Watanabe, Y, Omura, T, Cheng, R.H, Tsukihara, T. | | Deposit date: | 2003-05-23 | | Release date: | 2003-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The atomic structure of rice dwarf virus reveals the self-assembly mechanism of component proteins.
Structure, 11, 2003
|
|
4UIP
 
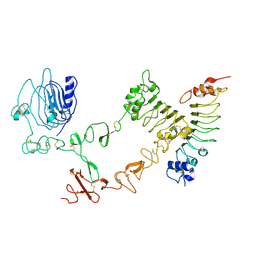 | | The complex structure of extracellular domain of EGFR with Repebody (rAC1). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Kang, Y.J, Cha, Y.J, Cho, H.S, Lee, J.J, Kim, H.S. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Prenylation and Oxime Ligation for the Synthesis of Stable and Homogeneous Protein-Drug Conjugates for Targeted Therapy.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6ISA
 
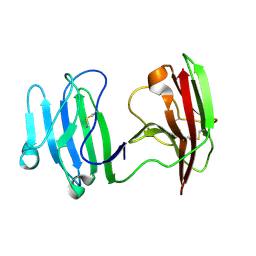 | | mCD226 | | Descriptor: | CD226 antigen | | Authors: | Wang, H, Qi, J, Zhang, S, Li, Y, Tan, S, Gao, G.F. | | Deposit date: | 2018-11-16 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding mode of the side-by-side two-IgV molecule CD226/DNAM-1 to its ligand CD155/Necl-5.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7PHR
 
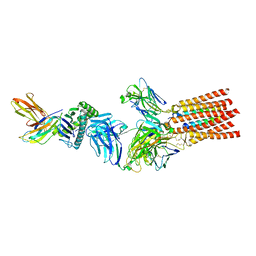 | | Structure of a fully assembled T-cell receptor engaging a tumor-associated peptide-MHC I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Susac, L, Thomas, C, Tampe, R. | | Deposit date: | 2021-08-18 | | Release date: | 2022-08-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC.
Cell, 185, 2022
|
|
