1KXX
 
 | |
1KOC
 
 | | RNA APTAMER COMPLEXED WITH ARGININE, NMR | | Descriptor: | ARGININE, RNA (5'-R(P*AP*CP*AP*GP*GP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*CP*GP*U)-3') | | Authors: | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | Deposit date: | 1996-03-28 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
7RZJ
 
 | | CRYSTAL STRUCTURE OF HLA-B*07:02 IN COMPLEX WITH MLL(747-755) PHOSPHOPEPTIDE | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-7 alpha chain, ... | | Authors: | Patskovsky, Y, Patskovska, L, Nyovanie, S, Natarajan, A, Joshi, B, Morin, B, Brittsan, C, Huber, O, Gordon, S, Michelet, X, Schmitzberger, F, Stein, R, Findeis, M, Hurwitz, A, Van Dijk, M, Buell, J, Underwood, D, Krogsgaard, M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular mechanism of phosphopeptide neoantigen immunogenicity.
Nat Commun, 14, 2023
|
|
1KVU
 
 | | UDP-GALACTOSE 4-EPIMERASE COMPLEXED WITH UDP-PHENOL | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Gulick, A.M, Holden, H.M. | | Deposit date: | 1997-03-07 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli.
Biochemistry, 36, 1997
|
|
1KUM
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KXE
 
 | |
5IGH
 
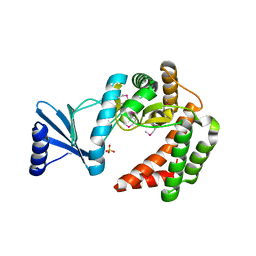 | | Macrolide 2'-phosphotransferase type I | | Descriptor: | Macrolide 2'-phosphotransferase, SULFATE ION | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2016-02-28 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Basis for Kinase-Mediated Macrolide Antibiotic Resistance.
Structure, 25, 2017
|
|
1KBS
 
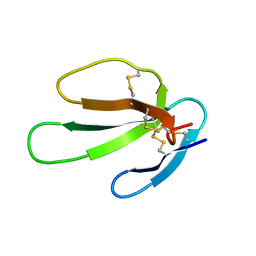 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 1 STRUCTURE | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
5IGV
 
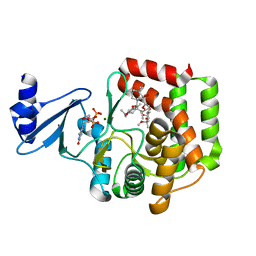 | |
1KBT
 
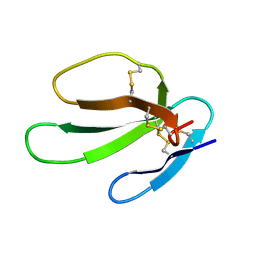 | | SOLUTION STRUCTURE OF CARDIOTOXIN IV, NMR, 12 STRUCTURES | | Descriptor: | CTX IV | | Authors: | Jeng, J.Y, Kumar, T.K.S, Jayaraman, G, Yu, C. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparison of the hemolytic activity and solution structures of two snake venom cardiotoxin analogues which only differ in their N-terminal amino acid.
Biochemistry, 36, 1997
|
|
1KPD
 
 | |
1KUL
 
 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KRT
 
 | |
1KEV
 
 | | STRUCTURE OF NADP-DEPENDENT ALCOHOL DEHYDROGENASE | | Descriptor: | NADP-DEPENDENT ALCOHOL DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Korkhin, Y, Frolow, F. | | Deposit date: | 1996-10-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystalline alcohol dehydrogenases from the mesophilic bacterium Clostridium beijerinckii and the thermophilic bacterium Thermoanaerobium brockii: preparation, characterization and molecular symmetry.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1KAA
 
 | |
1KAO
 
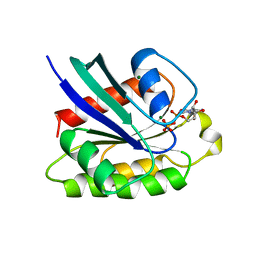 | | CRYSTAL STRUCTURE OF THE SMALL G PROTEIN RAP2A WITH GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RAP2A | | Authors: | Cherfils, J, Menetrey, J, Le Bras, G. | | Deposit date: | 1997-08-01 | | Release date: | 1997-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the small G protein Rap2A in complex with its substrate GTP, with GDP and with GTPgammaS.
EMBO J., 16, 1997
|
|
1KUN
 
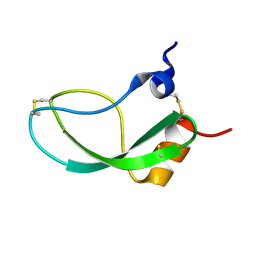 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | ALPHA3-CHAIN TYPE VI COLLAGEN | | Authors: | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-11-12 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
8ET5
 
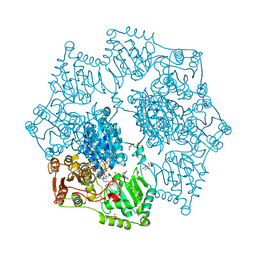 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase S653T mutant in complex with amidosulfuron | | Descriptor: | 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Acetolactate synthase, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-10-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal Structure of the Commercial Herbicide, Amidosulfuron, in Complex with Arabidopsis thaliana Acetohydroxyacid Synthase.
J.Agric.Food Chem., 71, 2023
|
|
5ILP
 
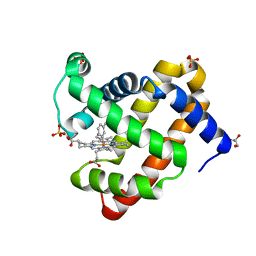 | | H64Q sperm whale myoglobin with a Fe-tolyl moiety | | Descriptor: | GLYCEROL, Myoglobin, SULFATE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
1KMN
 
 | |
1KAW
 
 | | STRUCTURE OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) | | Descriptor: | SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Raghunathan, S, Waksman, G. | | Deposit date: | 1996-12-06 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
8F86
 
 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
1KAP
 
 | | THREE-DIMENSIONAL STRUCTURE OF THE ALKALINE PROTEASE OF PSEUDOMONAS AERUGINOSA: A TWO-DOMAIN PROTEIN WITH A CALCIUM BINDING PARALLEL BETA ROLL MOTIF | | Descriptor: | ALKALINE PROTEASE, CALCIUM ION, TETRAPEPTIDE (GLY SER ASN SER), ... | | Authors: | Baumann, U, Wu, S, Flaherty, K.M, Mckay, D.B. | | Deposit date: | 1995-06-08 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: a two-domain protein with a calcium binding parallel beta roll motif.
EMBO J., 12, 1993
|
|
8F76
 
 | | Human olfactory receptor OR51E2 bound to propionate in complex with miniGs399 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Billesboelle, C.B, Manglik, A. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of odorant recognition by a human odorant receptor.
Nature, 615, 2023
|
|
1KDC
 
 | |
