301D
 
 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | Descriptor: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
2M27
 
 | | Major G-quadruplex structure formed in human VEGF promoter, a monomeric parallel-stranded quadruplex | | Descriptor: | DNA_(5'-D(*CP*GP*GP*GP*GP*CP*GP*GP*GP*CP*CP*TP*TP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3')_ | | Authors: | Agrawal, P, Hatzakis, E, Guo, K, Carver, M, Yang, D. | | Deposit date: | 2012-12-14 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes.
Nucleic Acids Res., 41, 2013
|
|
2MB4
 
 | | Solution structure of a stacked dimeric G-quadruplex formed by a segment of the human CEB1 minisatellite | | Descriptor: | DNA_(5'-D(*AP*GP*GP*GP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*TP*GP*G)-3') | | Authors: | Adrian, M, Ang, D.J, Lech, C, Heddi, B, Nicolas, A, Phan, A.T. | | Deposit date: | 2013-07-25 | | Release date: | 2014-05-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Conformational Dynamics of a Stacked Dimeric G-Quadruplex Formed by the Human CEB1 Minisatellite.
J.Am.Chem.Soc., 136, 2014
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Y
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
6GDS
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | Telomeric DNA (5' CTAACCCTAA) 10mer, Telomeric DNA (5'-TTAGGGTTAG)-3') 10mer | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6GDN
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | MAGNESIUM ION, Telomere DNA (42-MER) | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-24 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
6GDH
 
 | | Holliday Junctions formed from Telomeric DNA | | Descriptor: | DNA (5'-D(*CP*TP*AP*AP*CP*CP*CP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*GP*TP*TP*AP*G)-3') | | Authors: | Parkinson, G.N, Haider, S, Li, P, Khiali, S, Munnur, D, Ramanathan, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Holliday Junctions Formed from Human Telomeric DNA.
J. Am. Chem. Soc., 140, 2018
|
|
2BBR
 
 | | Crystal Structure of MC159 Reveals Molecular Mechanism of DISC Assembly and vFLIP Inhibition | | Descriptor: | AZIDE ION, Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Yang, J.K, Wang, L, Zheng, L, Wan, F, Ahmed, M, Lenardo, M.J, Wu, H. | | Deposit date: | 2005-10-17 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of MC159 reveals molecular mechanism of DISC assembly and FLIP inhibition.
Mol.Cell, 20, 2005
|
|
3QVT
 
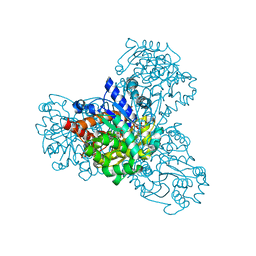 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus wild-type with the intermediate 5-keto 1-phospho glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
300D
 
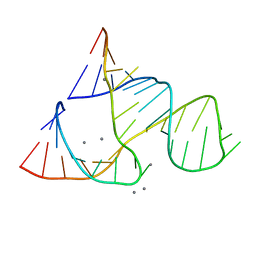 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
1R6F
 
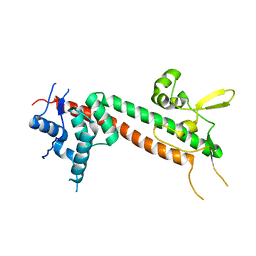 | | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague | | Descriptor: | Virulence-associated V antigen | | Authors: | Derewenda, U, Mateja, A, Devedjiev, Y, Routzahn, K.M, Evdokimov, A.G, Derewenda, Z.S, Waugh, D.S. | | Deposit date: | 2003-10-15 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague
Structure, 12, 2004
|
|
5U6A
 
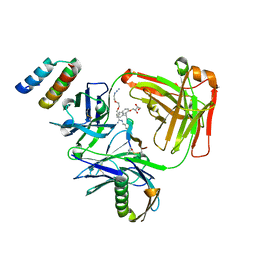 | | CRYSTAL STRUCTURE OF I83E MEDITOPE-ENABLED TRASTUZUMAB WITH AZIDO-PEG3-MEDITOPE | | Descriptor: | Heavy Chain, Immunoglobulin G binding protein A, Light Chain, ... | | Authors: | Williams, J.C, Bzymek, K.P, Pucket, J, Avery, K.A, Ma, Y, Xie, J, Zer, C, Horne, D. | | Deposit date: | 2016-12-07 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Crystal Structure Of I83E Meditope-Enabled Trastuzumab With Azido-PEG3-Meditope
To Be Published
|
|
6GHV
 
 | | Structure of a DC-SIGN CRD in complex with high affinity glycomimetic. | | Descriptor: | CALCIUM ION, CD209 antigen, CHLORIDE ION, ... | | Authors: | Thepaut, M, Achilli, S, Medve, L, Bernardi, A, Fieschi, F. | | Deposit date: | 2018-05-09 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enhancing Potency and Selectivity of a DC-SIGN Glycomimetic Ligand by Fragment-Based Design: Structural Basis.
Chemistry, 25, 2019
|
|
3E9J
 
 | |
4OOW
 
 | | HCV NS5B polymerase with a fragment of quercetagetin | | Descriptor: | CATECHOL, RNA-directed RNA polymerase | | Authors: | Guichou, J.F, Ahmed-Belkacem, A, Rozenn, B, Nazim, N, Hernandez, E, Pallier, C, Pawlotsky, J.M. | | Deposit date: | 2014-02-04 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention.
Nucleic Acids Res., 42, 2014
|
|
2W53
 
 | | Structure of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. | | Descriptor: | REPRESSOR, SULFATE ION | | Authors: | Mate, M.J, Romero, A, Hernandez, A, Martinez, J.L. | | Deposit date: | 2008-12-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Smet, the Repressor of the Stenotrophomonas Maltophilia Multidrug Efflux Pump Smedef.
J.Biol.Chem., 284, 2009
|
|
3MVN
 
 | | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP | | Descriptor: | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, F.W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-04 | | Release date: | 2010-06-16 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a domain from a putative UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-medo-diaminopimelate ligase from Haemophilus ducreyi 35000HP
To be Published
|
|
2GA3
 
 | |
3GDJ
 
 | |
2YKA
 
 | | RRM domain of mRNA export adaptor REF2-I bound to HVS ORF57 peptide | | Descriptor: | 52 KDA IMMEDIATE-EARLY PHOSPHOPROTEIN, RNA AND EXPORT FACTOR-BINDING PROTEIN 2 | | Authors: | Tunnicliffe, R.B, Hautbergue, G.M, Kalra, P, Wilson, S.A, Golovanov, A.P. | | Deposit date: | 2011-05-26 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri Orf57 to the Mammalian Export Adaptor Alyref.
Plos Pathog., 10, 2014
|
|
4NXI
 
 | | Rv2466c Mediates the Activation of TP053 To Kill Replicating and Non-replicating Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Uncharacterized protein | | Authors: | Albesa-Jove, D, Urresti, S, Comino, N, Guerin, M.E. | | Deposit date: | 2013-12-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Rv2466c mediates the activation of TP053 to kill replicating and non-replicating Mycobacterium tuberculosis.
Acs Chem.Biol., 9, 2014
|
|
