3QY1
 
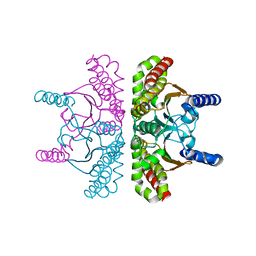 | | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-03-02 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | 1.54A Resolution Crystal Structure of a Beta-Carbonic Anhydrase from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
5ZO1
 
 | | Crystal structure of mouse nectin-like molecule 4 (mNecl-4) full ectodomain (Ig1-Ig3), 2.2A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Cell adhesion molecule 4, GLYCEROL | | Authors: | Liu, X, An, T, Li, D, Fan, Z, Xiang, P, Li, C, Ju, W, Li, J, Hu, G, Qin, B, Yin, B, Wojdyla, J.A, Wang, M, Yuan, J, Qiang, B, Shu, P, Cui, S, Peng, X. | | Deposit date: | 2018-04-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6HZN
 
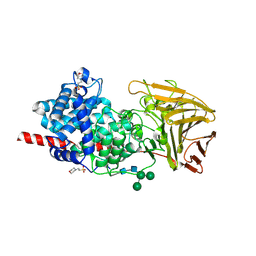 | | Crystal structure of human dermatan sulfate epimerase 1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hasan, M, Unge, J, Westergren-Thorsson, G, Ellervik, U, Mueller, U, Malmstrom, A, Tykesson, E. | | Deposit date: | 2018-10-23 | | Release date: | 2020-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The structure of human dermatan sulfate epimerase 1 emphasizes the importance of C5-epimerization of glucuronic acid in higher organisms
Chem Sci, 2020
|
|
1X0V
 
 | |
7CP5
 
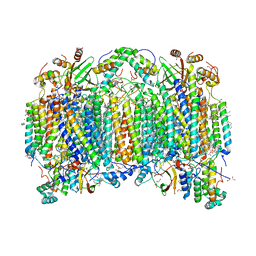 | | Bovine heart cytochrome c oxidase in a catalytic intermediate of E at 1.76 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Tsukihara, T, Shimada, A. | | Deposit date: | 2020-08-06 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Critical roles of the Cu B site in efficient proton pumping as revealed by crystal structures of mammalian cytochrome c oxidase catalytic intermediates.
J.Biol.Chem., 297, 2021
|
|
3KKI
 
 | | PLP-Dependent Acyl-CoA transferase CqsA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CAI-1 autoinducer synthase, MAGNESIUM ION, ... | | Authors: | Kelly, R.C, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio cholerae quorum-sensing autoinducer CAI-1: analysis of the biosynthetic enzyme CqsA.
Nat.Chem.Biol., 5, 2009
|
|
5ELW
 
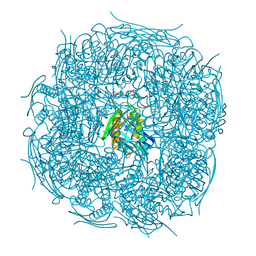 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (R)-C348, to 1.36A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WV3
 
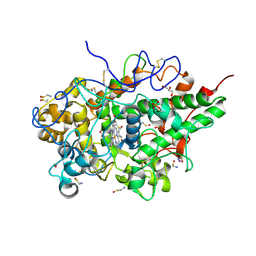 | | Crystal structure of bovine lactoperoxidase with a partial Glu258-heme linkage at 2.07 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-12-21 | | Release date: | 2017-02-15 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis of activation of mammalian heme peroxidases
Prog. Biophys. Mol. Biol., 133, 2018
|
|
4KAK
 
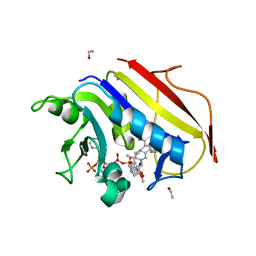 | | Crystal structure of human dihydrofolate reductase complexed with NADPH and 6-ethyl-5-[(3S)-3-[3-methoxy-5-(pyridine-4-yl)phenyl]but-1-yn-1-yl]pyrimidine-2,4-diamine (UCP1006) | | Descriptor: | 6-ethyl-5-{(3R)-3-[3-methoxy-5-(pyridin-4-yl)phenyl]but-1-yn-1-yl}pyrimidine-2,4-diamine, CALCIUM ION, Dihydrofolate reductase, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-22 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
2AQH
 
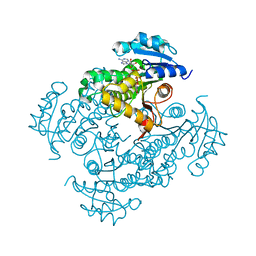 | | Crystal structure of Isoniazid-resistant I21V Enoyl-ACP(CoA) reductase mutant enzyme from Mycobacterium tuberculosis in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Oliveira, J.S, Pereira, J.H, Rodrigues, N.C, Canduri, F, de Souza, O.N, Basso, L.A, de Azevedo Jr, W.F, Santos, D.S. | | Deposit date: | 2005-08-18 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystallographic and Pre-steady-state Kinetics Studies on Binding of NADH to Wild-type and Isoniazid-resistant Enoyl-ACP(CoA) Reductase Enzymes from Mycobacterium tuberculosis.
J.Mol.Biol., 359, 2006
|
|
4D74
 
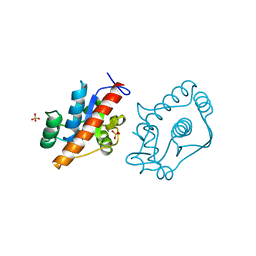 | | 1.57 A crystal structure of erwinia amylovora tyrosine phosphatase amsI | | Descriptor: | PROTEIN-TYROSINE-PHOSPHATASE AMSI, SULFATE ION | | Authors: | Benini, S, Salomone-Stagni, M, Caputi, L, Cianci, M. | | Deposit date: | 2014-11-19 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Characterization and 1.57 A Resolution Structure of the Key Fire Blight Phosphatase Amsi from Erwinia Amylovora
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1Y7T
 
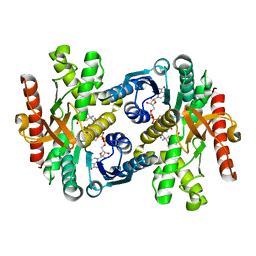 | | Crystal structure of NAD(H)-depenent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
7YUY
 
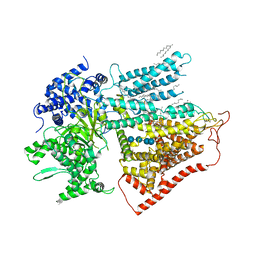 | | Structure of a mutated membrane-bound glycosyltransferase | | Descriptor: | (11R,14S)-17-amino-14-hydroxy-8,14-dioxo-9,13,15-trioxa-14lambda~5~-phosphaheptadecan-11-yl decanoate, 1,3-beta-glucan synthase component FKS1, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, X.L, Yang, P, Zhang, M, Liu, X.T, Yu, H.J. | | Deposit date: | 2022-08-18 | | Release date: | 2023-03-29 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and mechanistic insights into fungal beta-1,3-glucan synthase FKS1.
Nature, 616, 2023
|
|
1NR0
 
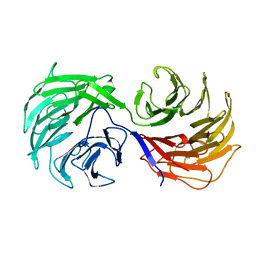 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | Descriptor: | Actin interacting protein 1, MANGANESE (II) ION | | Authors: | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
4GUC
 
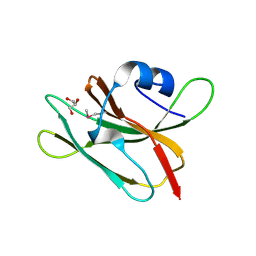 | | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Protein BA_2500, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom resolution crystal structure of uncharacterized protein BA_2500 from Bacillus anthracis str. Ames
To be Published
|
|
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
1Z5Y
 
 | | Crystal Structure Of The Disulfide-Linked Complex Between The N-Terminal Domain Of The Electron Transfer Catalyst DsbD and The Cytochrome c Biogenesis Protein CcmG | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Thiol:disulfide interchange protein dsbD, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Gruetter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2005-03-21 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural Basis and Kinetics of DsbD-Dependent Cytochrome c Maturation
STRUCTURE, 13, 2005
|
|
4H3D
 
 | | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid. | | Descriptor: | 3-dehydroquinate dehydratase, 5-hydroxy-6-methyl-4-oxo-4H-pyran-2-carboxylic acid, ACETATE ION, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Duban, M.-E, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of of Type I 3-Dehydroquinate Dehydratase (aroD) from Clostridium difficile with Covalent Modified Comenic Acid.
TO BE PUBLISHED
|
|
5ZF0
 
 | | X-ray Structure of the Electron Transfer Complex between Ferredoxin and Photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kubota-Kawai, H, Mutoh, R, Shinmura, K, Setif, P, Nowaczyk, M, Roegner, M, Ikegami, T, Tanaka, T, Kurisu, G. | | Deposit date: | 2018-03-01 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | X-ray structure of an asymmetrical trimeric ferredoxin-photosystem I complex
Nat Plants, 4, 2018
|
|
5Z06
 
 | | Crystal structure of beta-1,2-glucanase from Parabacteroides distasonis | | Descriptor: | BDI_3064 protein, CALCIUM ION, GLYCEROL | | Authors: | Shimizu, H, Nakajima, M, Miyanaga, A, Takahashi, Y, Tanaka, N, Kobayashi, K, Sugimoto, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-12-18 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and Structural Analysis of a Novel exo-Type Enzyme Acting on beta-1,2-Glucooligosaccharides from Parabacteroides distasonis
Biochemistry, 57, 2018
|
|
1ZGK
 
 | | 1.35 angstrom structure of the Kelch domain of Keap1 | | Descriptor: | Kelch-like ECH-associated protein 1 | | Authors: | Li, X, Bottoms, C.A, Hannink, M, Beamer, L.J. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Conserved solvent and side-chain interactions in the 1.35 Angstrom structure of the Kelch domain of Keap1.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
3UVE
 
 | |
4KD7
 
 | | Human dihydrofolate reductase complexed with NADPH and 5-{3-[3-methoxy-5(pyridine-4-yl)phenyl]prop-1-yn-1-yl}-6-ethyl-pyrimidine-2,4-diamine | | Descriptor: | 6-ethyl-5-{3-[3-methoxy-5-(pyridin-4-yl)phenyl]prop-1-yn-1-yl}pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lamb, K.M, Anderson, A.C. | | Deposit date: | 2013-04-24 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.715 Å) | | Cite: | Elucidating features that drive the design of selective antifolates using crystal structures of human dihydrofolate reductase.
Biochemistry, 52, 2013
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | Authors: | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | Deposit date: | 2018-12-27 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
