2UXS
 
 | | 2.7A crystal structure of inorganic pyrophosphatase (Rv3628) from Mycobacterium tuberculosis at pH 7.5 | | Descriptor: | INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Cole, R.E, Cianci, M, Hall, J.F, Matsuda, T, Kigawa, T, Yokoyama, S, Hasnain, S.S, Tabernero, L. | | Deposit date: | 2007-03-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Rv3628: An Inorganic Pyrophosphatase from Mycobacterium Tuberculosis
To be Published
|
|
2UYF
 
 | | Single mutant F111L DntR from Burkholderia sp. strain DNT in complex with thiocyanate | | Descriptor: | ACETATE ION, GLYCEROL, REGULATORY PROTEIN, ... | | Authors: | Lonneborg, R, Smirova, I, Dian, C, Leonard, G.A, Mcsweeney, S, Brzezinski, P. | | Deposit date: | 2007-04-04 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Vivo and in Vitro Investigation of Transcriptional Regulation by Dntr.
J.Mol.Biol., 372, 2007
|
|
4EEG
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-Gal-Beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
1H2A
 
 | | SINGLE CRYSTALS OF HYDROGENASE FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FE3-S4 CLUSTER, HYDROGENASE, IRON/SULFUR CLUSTER, ... | | Authors: | Higuchi, Y, Yasuoka, N. | | Deposit date: | 1997-10-17 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis.
Structure, 5, 1997
|
|
4EEA
 
 | | Crystal structure of human M340H-beta-1,4-galactosyltransferase-1 (M340H-B4GAL-T1) in complex with GLCNAC-BETA1,6-Gal-Beta1,4-Glc-BETA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Beta-1,4-galactosyltransferase 1, GLYCEROL, ... | | Authors: | Ramakrishnan, B, Qasba, P.K. | | Deposit date: | 2012-03-28 | | Release date: | 2012-07-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding of N-acetylglucosamine (GlcNAc) beta 1-6-branched oligosaccharide acceptors to beta 4-galactosyltransferase I reveals a new ligand binding mode.
J.Biol.Chem., 287, 2012
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | Descriptor: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6FTZ
 
 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
2II5
 
 | | Crystal structure of a cubic core of the dihydrolipoamide acyltransferase (E2b) component in the branched-chain alpha-ketoacid dehydrogenase complex (BCKDC), Isobutyryl-Coenzyme A-bound form | | Descriptor: | ACETATE ION, CHLORIDE ION, ISOBUTYRYL-COENZYME A, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Brautigam, C.A, Custorio, M, Chuang, D.T. | | Deposit date: | 2006-09-27 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synchronized substrate-gating mechanism revealed by cubic-core structure of the bovine branched-chain alpha-ketoacid dehydrogenase complex.
Embo J., 25, 2006
|
|
7CUO
 
 | | IclR transcription factor complexed with 4-hydroxybenzoic acid from Microbacterium hydrocarbonoxydans | | Descriptor: | P-HYDROXYBENZOIC ACID, SULFATE ION, Transcription factor | | Authors: | Akiyama, T, Ito, S, Sasaki, Y, Yajima, S. | | Deposit date: | 2020-08-23 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the conformational changes in Microbacterium hydrocarbonoxydans IclR transcription factor homolog due to ligand binding.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
4EY4
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in the Apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
4EY7
 
 | | Crystal Structure of Recombinant Human Acetylcholinesterase in Complex with Donepezil | | Descriptor: | 1,2-ETHANEDIOL, 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cheung, J, Rudolph, M, Burshteyn, F, Cassidy, M, Gary, E, Love, J, Height, J, Franklin, M. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3509 Å) | | Cite: | Structures of human acetylcholinesterase in complex with pharmacologically important ligands.
J.Med.Chem., 55, 2012
|
|
4V4E
 
 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici complexed with inhibitor 1,2,4,5-tetrahydroxy-benzene | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, BENZENE-1,2,4,5-TETROL, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3I6O
 
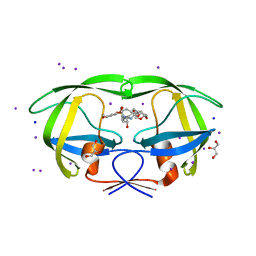 | | Crystal structure of wild type HIV-1 protease with macrocyclic inhibitor GRL-0216A | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-2-hydroxy-3-[(7E)-13-methoxy-1,1-dioxido-3,4,5,6,9,10-hexahydro-2H-11,1,2-benzoxathiazacyclotridecin-2-yl]propyl}carbamate, GLYCEROL, IODIDE ION, ... | | Authors: | Chumanevich, A.A, Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
3DTV
 
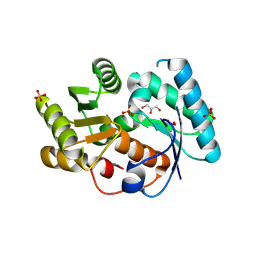 | | Crystal structure of arylmalonate decarboxylase | | Descriptor: | Arylmalonate decarboxylase, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Nakasako, M, Obata, R, Miyamaoto, K, Ohta, H. | | Deposit date: | 2008-07-16 | | Release date: | 2009-07-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Inverting the Enantioselectivity of Arylmalonate Decarboxylase Revealed by the Structural Analysis of the Gly74Cys/Cys188Ser Mutant in the Liganded Form
Biochemistry, 49, 2010
|
|
4V4C
 
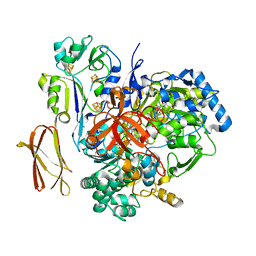 | | Crystal Structure of Pyrogallol-Phloroglucinol Transhydroxylase from Pelobacter acidigallici | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, ACETATE ION, CALCIUM ION, ... | | Authors: | Messerschmidt, A, Niessen, H, Abt, D, Einsle, O, Schink, B, Kroneck, P.M.H. | | Deposit date: | 2004-06-02 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
3B6D
 
 | |
1S1M
 
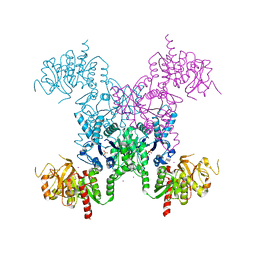 | | Crystal Structure of E. Coli CTP Synthetase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CTP synthase, IODIDE ION, ... | | Authors: | Endrizzi, J.A, Kim, H, Anderson, P.M, Baldwin, E.P. | | Deposit date: | 2004-01-06 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Escherichia coli Cytidine Triphosphate Synthetase, a Nucleotide-Regulated Glutamine Amidotransferase/ATP-Dependent Amidoligase Fusion Protein and Homologue of Anticancer and Antiparasitic Drug Targets
Biochemistry, 43, 2004
|
|
5HWC
 
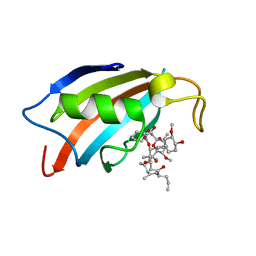 | |
1D2J
 
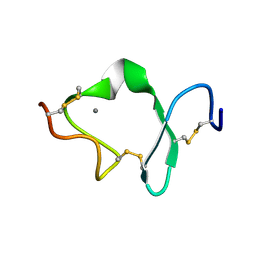 | | LDL RECEPTOR LIGAND-BINDING MODULE 6 | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR | | Authors: | North, C.L, Blacklow, S.C. | | Deposit date: | 1999-09-23 | | Release date: | 2000-03-22 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the sixth LDL-A module of the LDL receptor.
Biochemistry, 39, 2000
|
|
8OLT
 
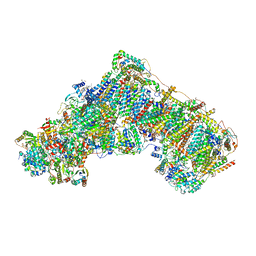 | | Mitochondrial complex I from Mus musculus in the active state bound with piericidin A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-30 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
8OM1
 
 | | Mitochondrial complex I from Mus musculus in the active state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Grba, D.N, Chung, I, Bridges, H.R, Agip, A.N.A, Hirst, J. | | Deposit date: | 2023-03-31 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Investigation of hydrated channels and proton pathways in a high-resolution cryo-EM structure of mammalian complex I.
Sci Adv, 9, 2023
|
|
6FTY
 
 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 5 | | Descriptor: | 4-[[(5~{S},7~{R})-3-azanyl-1-adamantyl]carbonylamino]-1~{H}-indole-2-carboxamide, Complement factor D, GLYCEROL | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
4A8V
 
 | | Crystal Structure of Birch Pollen Allergen Bet v 1 isoform j in complex with 8-Anilinonaphthalene-1-sulfonate (ANS) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, ... | | Authors: | Kofler, S, Brandstetter, H. | | Deposit date: | 2011-11-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Crystallographically Mapped Ligand Binding Differs in High and Low Ige Binding Isoforms of Birch Pollen Allergen Bet V 1.
J.Mol.Biol., 422, 2012
|
|
6A7I
 
 | | CYP154C4 from Streptomyces sp. W2061 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lee, C.W, Lee, J.H. | | Deposit date: | 2018-07-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Characterization of two steroid hydroxylases from different Streptomyces spp. and their ligand-bound and -unbound crystal structures.
Febs J., 286, 2019
|
|
3EDU
 
 | | Crystal structure of the ankyrin-binding domain of human erythroid spectrin | | Descriptor: | Spectrin beta chain, erythrocyte | | Authors: | Simonovic, M, Stabach, P, Simonovic, I, Steitz, T.A, Morrow, J.S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the ankyrin-binding site of {beta}-spectrin reveals how tandem spectrin-repeats generate unique ligand-binding properties
Blood, 113, 2009
|
|
