1FHZ
 
 | | PSORALEN CROSS-LINKED D(CCGGTACCGG) FORMS HOLLIDAY JUNCTION | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*CP*CP*GP*GP*TP*AP*CP*CP*GP*G)-3') | | Authors: | Eichman, B.F, Mooers, B.H.M, Alberti, M, Hearst, J.E, Ho, P.S. | | Deposit date: | 2000-08-02 | | Release date: | 2001-04-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of psoralen cross-linked DNAs: drug-dependent formation of Holliday junctions.
J.Mol.Biol., 308, 2001
|
|
2NTZ
 
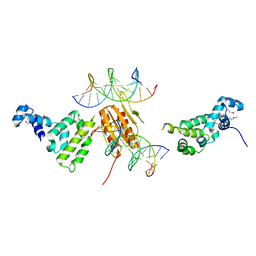 | |
1RRS
 
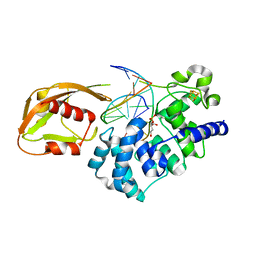 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
4BXO
 
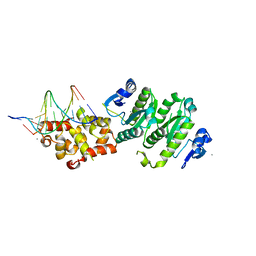 | | Architecture and DNA recognition elements of the Fanconi anemia FANCM- FAAP24 complex | | Descriptor: | 5'-D(*GP*AP*TP*GP*AP*TP*GP*CP*TP*GP*CP)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*AP*TP*CP)-3', CALCIUM ION, ... | | Authors: | Coulthard, R, Deans, A, Swuec, P, Bowles, M, Purkiss, A, Costa, A, West, S, McDonald, N. | | Deposit date: | 2013-07-15 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Architecture and DNA Recognition Elements of the Fanconi Anemia Fancm-Faap24 Complex.
Structure, 21, 2013
|
|
1SA3
 
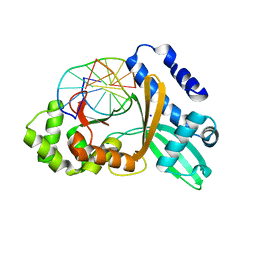 | | An asymmetric complex of restriction endonuclease MspI on its palindromic DNA recognition site | | Descriptor: | 5'-D(*CP*CP*CP*CP*CP*GP*GP*GP*GP*G)-3', SODIUM ION, Type II restriction enzyme MspI | | Authors: | Xu, Q.S, Kucera, R.B, Roberts, R.J, Guo, H.C. | | Deposit date: | 2004-02-06 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An Asymmetric Complex of Restriction Endonuclease MspI on Its Palindromic DNA Recognition Site.
STRUCTURE, 12, 2004
|
|
3WI3
 
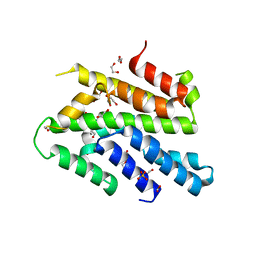 | | Crystal Structure of the Sld3/Treslin domain from yeast Sld3 | | Descriptor: | 1,2-ETHANEDIOL, DNA replication regulator SLD3, SULFATE ION | | Authors: | Itou, H, Araki, H, Shirakihara, Y. | | Deposit date: | 2013-09-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the homology domain of the eukaryotic DNA replication proteins sld3/treslin.
Structure, 22, 2014
|
|
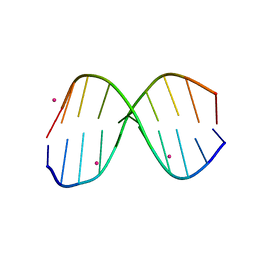
3R86
 
 | |
1NAB
 
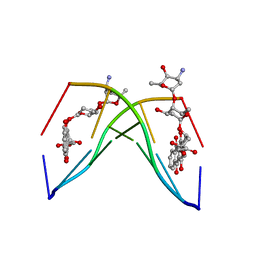 | | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', 7-[5-(4-AMINO-5-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY)-4-HYDROXY-6-METHYL-TETRAHYDRO-PYRAN-2-YLOXY]-6,9,11-TRIHYDROXY-9-(2-HYDROXY-ACETYL)-7,8,9,10-TETRAHYDRO-NAPHTHACENE-5,12-DIONE | | Authors: | Temperini, C, Messori, L, Orioli, P, Di Bugno, C, Animati, F, Ughetto, G. | | Deposit date: | 2002-11-27 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the complex between a disaccharide anthracycline and the DNA hexamer d(CGATCG) reveals two different binding sites involving two DNA duplexes
Nucleic Acids Res., 31, 2003
|
|
1P20
 
 | |
6P66
 
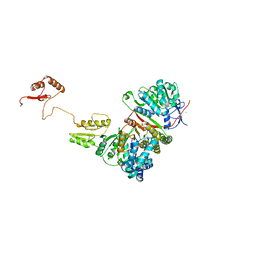 | |
1QTW
 
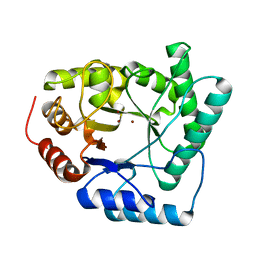 | | HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI DNA REPAIR ENZYME ENDONUCLEASE IV | | Descriptor: | ENDONUCLEASE IV, ZINC ION | | Authors: | Hosfield, D.J, Guan, Y, Haas, B.J, Cunningham, R.P, Tainer, J.A. | | Deposit date: | 1999-06-29 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis.
Cell(Cambridge,Mass.), 98, 1999
|
|
1DPU
 
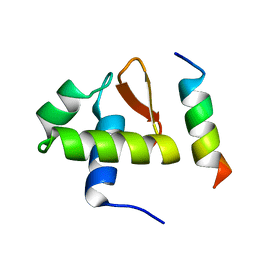 | | SOLUTION STRUCTURE OF THE C-TERMINAL DOMAIN OF HUMAN RPA32 COMPLEXED WITH UNG2(73-88) | | Descriptor: | REPLICATION PROTEIN A (RPA32) C-TERMINAL DOMAIN, URACIL DNA GLYCOSYLASE (UNG2) | | Authors: | Mer, G, Edwards, A.M, Chazin, W.J. | | Deposit date: | 1999-12-27 | | Release date: | 2000-11-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA.
Cell(Cambridge,Mass.), 103, 2000
|
|
6G01
 
 | |
3I2O
 
 | |
3I3M
 
 | |
404D
 
 | |
2VN2
 
 | | Crystal structure of the N-terminal domain of DnaD protein from Geobacillus kaustophilus HTA426 | | Descriptor: | CHROMOSOME REPLICATION INITIATION PROTEIN, MAGNESIUM ION | | Authors: | Huang, C.-Y, Chang, Y.-W, Chen, W.-T, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2008-01-30 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the N-Terminal Domain of Geobacillus Kaustophilus Hta426 Dnad Protein.
Biochem.Biophys.Res.Commun., 375, 2008
|
|
2JOX
 
 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
6G02
 
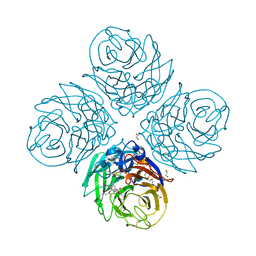 | | Complex of neuraminidase from H1N1 influenza virus with tamiphosphor omega-azidohexyl ester | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-[[(3~{R},4~{R},5~{S})-4-acetamido-5-azanyl-3-pentan-3-yloxy-cyclohexen-1-yl]-oxidanyl-phosphoryl]oxyhexylimino-azanylidene-azanium, ... | | Authors: | Pachl, P, Pokorna, J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | DNA-linked inhibitor antibody assay (DIANA) as a new method for screening influenza neuraminidase inhibitors.
Biochem. J., 475, 2018
|
|
1PQQ
 
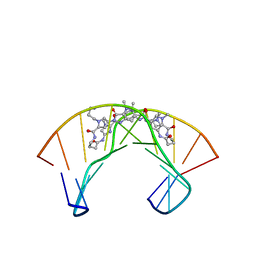 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
5ESP
 
 | |
5F9L
 
 | |
1Q3V
 
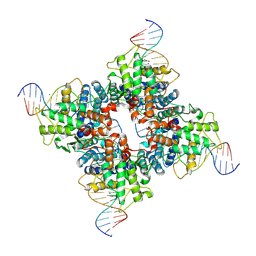 | | Crystal structure of a wild-type Cre recombinase-loxP synapse: phosphotyrosine covalent intermediate | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-08-01 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
1Z3F
 
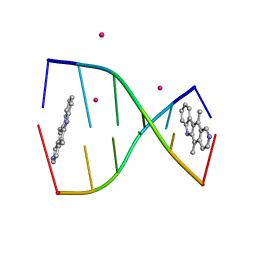 | | Structure of ellipticine in complex with a 6-bp DNA | | Descriptor: | 5'-D(*CP*GP*AP*TP*CP*G)-3', COBALT (II) ION, ELLIPTICINE | | Authors: | Canals, A, Purciolas, M, Aymami, J, Coll, M. | | Deposit date: | 2005-03-12 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The anticancer agent ellipticine unwinds DNA by intercalative binding in an orientation parallel to base pairs.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5F9N
 
 | |
