4QL7
 
 | | Crystal structure of C-terminus truncated Alkylhydroperoxide Reductase subunit C (AhpC1-172) from E. coli | | Descriptor: | Alkylhydroperoxide Reductase subunit C | | Authors: | Kamariah, N, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2014-06-11 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Key roles of the Escherichia coli AhpC C-terminus in assembly and catalysis of alkylhydroperoxide reductase, an enzyme essential for the alleviation of oxidative stress.
Biochim.Biophys.Acta, 1837, 2014
|
|
4QL9
 
 | |
1RJH
 
 | | Structure of the Calcium Free Form of the C-type Lectin-like Domain of Tetranectin | | Descriptor: | Tetranectin | | Authors: | Nielbo, S, Thomsen, J.K, Graversen, J.H, Etzerodt, M, Poulsen, F.M, Thoegersen, H.C. | | Deposit date: | 2003-11-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the Plasminogen Kringle 4 Binding Calcium-Free Form of the C-Type Lectin-Like Domain of Tetranectin.
Biochemistry, 43, 2004
|
|
6LBG
 
 | | Structure of OR51B2 bound FEM1C | | Descriptor: | Protein fem-1 homolog C,Peptide from Olfactory receptor 51B2, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
5TRG
 
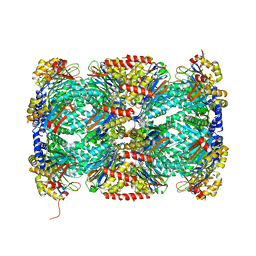 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
6LE6
 
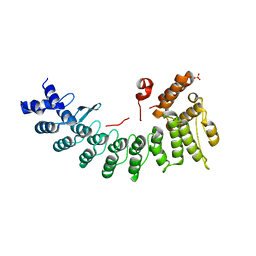 | | Structure of LNLPTQGRAR bound FEM1C | | Descriptor: | Protein fem-1 homolog C,10-mer peptide, SULFATE ION | | Authors: | Chen, X, Liao, S, Xu, C. | | Deposit date: | 2019-11-24 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Molecular basis for arginine C-terminal degron recognition by Cul2 FEM1 E3 ligase.
Nat.Chem.Biol., 17, 2021
|
|
2TN4
 
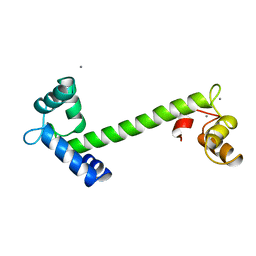 | | FOUR CALCIUM TNC | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Love, M.L, Dominguez, R, Houdusse, A, Cohen, C. | | Deposit date: | 1997-09-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of four Ca2+-bound troponin C at 2.0 A resolution: further insights into the Ca2+-switch in the calmodulin superfamily.
Structure, 5, 1997
|
|
3CJ0
 
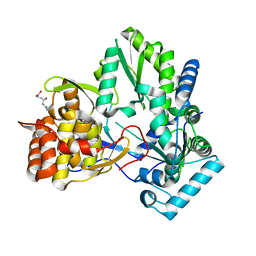 | |
1ERJ
 
 | | CRYSTAL STRUCTURE OF THE C-TERMINAL WD40 DOMAIN OF TUP1 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR TUP1 | | Authors: | Sprague, E.R, Redd, M.J, Johnson, A.D, Wolberger, C. | | Deposit date: | 2000-04-06 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast.
EMBO J., 19, 2000
|
|
2AX1
 
 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5ee) | | Descriptor: | 5R-(3,4-DICHLOROPHENYLMETHYL)-3-(2-THIOPHENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
3CJ3
 
 | |
3CJ2
 
 | |
2EDN
 
 | | Solution structure of the first ig-like domain from human Myosin-binding protein C, fast-type | | Descriptor: | Myosin-binding protein C, fast-type | | Authors: | Nagashima, K, Kurosaki, C, Yoshida, M, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-14 | | Release date: | 2007-08-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first ig-like domain from human Myosin-binding protein C, fast-type
To be Published
|
|
2VIF
 
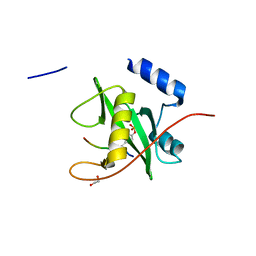 | | Crystal structure of SOCS6 SH2 domain in complex with a c-KIT phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, MAST/STEM CELL GROWTH FACTOR RECEPTOR, SUPPRESSOR OF CYTOKINE SIGNALLING 6 | | Authors: | Bullock, A, Pike, A.C.W, Savitsky, P, Keates, T, Pilka, E.S, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Knapp, S. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for C-Kit Inhibition by the Suppressor of Cytokine Signaling 6 (Socs6) Ubiquitin Ligase.
J.Biol.Chem., 286, 2011
|
|
7CLZ
 
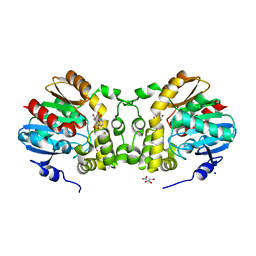 | |
1H4J
 
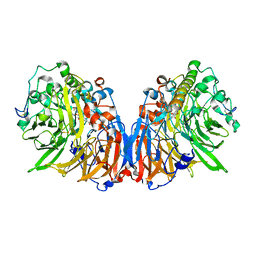 | | Methylobacterium extorquens methanol dehydrogenase D303E mutant | | Descriptor: | CALCIUM ION, Methanol dehydrogenase [cytochrome c] subunit 1, Methanol dehydrogenase [cytochrome c] subunit 2, ... | | Authors: | Mohammed, F, Gill, R, Thompson, D, Cooper, J.B, Wood, S.P, Afolabi, P.R, Anthony, C. | | Deposit date: | 2001-05-11 | | Release date: | 2001-08-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Site-Directed Mutagenesis and X-Ray Crystallography of the Pqq-Containing Quinoprotein Methanol Dehydrogenase and its Electron Acceptor, Cytochrome C(L)(,)
Biochemistry, 40, 2001
|
|
2QEE
 
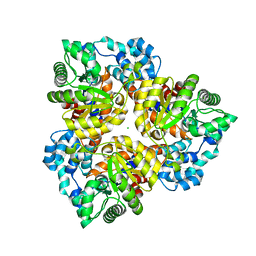 | | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125 | | Descriptor: | BH0493 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Malashkevich, V.N, Toro, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-06-25 | | Release date: | 2007-07-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putative amidohydrolase BH0493 from Bacillus halodurans C-125.
To be Published
|
|
2AWZ
 
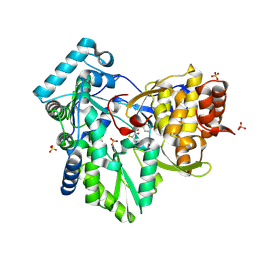 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
3QGH
 
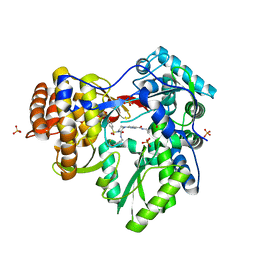 | |
3SS3
 
 | |
2JDL
 
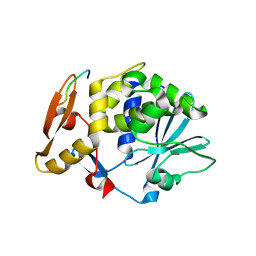 | | Structure of C-terminal region of acidic P2 ribosomal protein complexed with trichosanthin | | Descriptor: | ACIDIC RIBOSOMAL PROTEIN P2, RIBOSOME-INACTIVATING PROTEIN ALPHA-TRICHOSANTHIN | | Authors: | Too, P.H, Mak, A.N, Zhu, G, Au, S.W, Wong, K.B, Shaw, P.C. | | Deposit date: | 2007-01-11 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Nucleic Acids Res., 37, 2009
|
|
3SS5
 
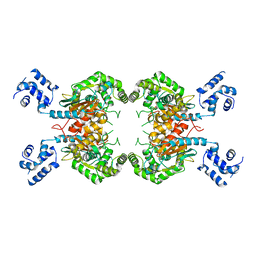 | |
1H3M
 
 | | Structure of 4-diphosphocytidyl-2C-methyl-D-erythritol synthetase | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLORIDE ION, PENTANE-1,5-DIAMINE | | Authors: | Kemp, L.E, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-09-10 | | Release date: | 2003-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Tetragonal Crystal Form of Escherichia Coli 2-C-Methyl-D-Erythritol 4-Phosphate Cytidylyltransferase
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3PYD
 
 | | crystal structure of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis | | Descriptor: | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase, GLYCEROL | | Authors: | Shan, S, Chen, X.H, Liu, T, Zhao, H.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | The Structural Basis for anti-TB Drug Discovery targeting of 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (IspE) from Mycobacterium tuberculosis
To be Published
|
|
1HEI
 
 | |
