7O1V
 
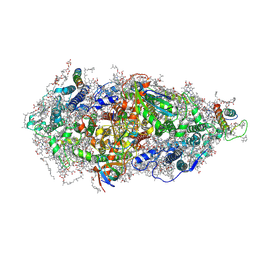 | | Structure of a Minimal Photosystem I | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Nelson, N, Caspy, I, Lambrev, P. | | Deposit date: | 2021-03-30 | | Release date: | 2021-09-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Two-Dimensional Electronic Spectroscopy of a Minimal Photosystem I Complex Reveals the Rate of Primary Charge Separation.
J.Am.Chem.Soc., 143, 2021
|
|
5KNU
 
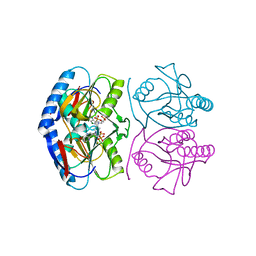 | | Crystal structure of E. coli hypoxanthine phosphoribosyltransferase in complexed with 9-[N,N-(Bis-3-phosphonopropyl)aminomethyl]-9-deazahypoxanthine | | Descriptor: | 3-[(4-oxidanylidene-3,5-dihydropyrrolo[3,2-d]pyrimidin-7-yl)methyl-(3-phosphonopropyl)amino]propylphosphonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypoxanthine-guanine phosphoribosyltransferase, ... | | Authors: | Eng, W.S, Keough, D.T, Baszczynski, O, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.808 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
8ERP
 
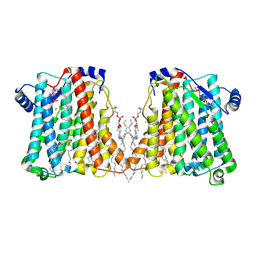 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP-choline | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Cholinephosphotransferase 1, MAGNESIUM ION, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
8ERO
 
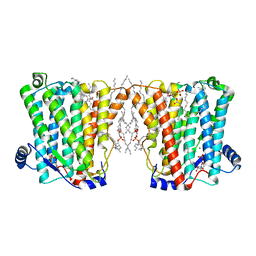 | | Structure of Xenopus cholinephosphotransferase1 in complex with CDP | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CYTIDINE-5'-DIPHOSPHATE, Cholinephosphotransferase 1, ... | | Authors: | Wang, L, Zhou, M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-04-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a eukaryotic cholinephosphotransferase-1 reveals mechanisms of substrate recognition and catalysis.
Nat Commun, 14, 2023
|
|
5KNV
 
 | | E coli hypoxanthine guanine phosphoribosyltransferase in complexed with 9-[(N-Phosphonoethyl-N-phosphonobutyl)-2-aminoethyl]-hypoxanthine | | Descriptor: | 2-[2-(6-oxidanylidene-1~{H}-purin-9-yl)ethyl-(4-phosphonobutyl)amino]ethylphosphonic acid, Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION | | Authors: | Eng, W.S, Keough, D.T, Hockova, D, Janeba, Z. | | Deposit date: | 2016-06-28 | | Release date: | 2017-07-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.861 Å) | | Cite: | Crystal Structures of Acyclic Nucleoside Phosphonates in Complex with Escherichia coli Hypoxanthine Phosphoribosyltransferase
Chemistryselect, 1, 2016
|
|
5L71
 
 | | Crystal structure of mouse phospholipid hydroperoxide glutathione peroxidase 4 (GPx4) | | Descriptor: | 1,2-ETHANEDIOL, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Janowski, R, Scanu, S, Madl, T, Niessing, D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal and solution structural studies of mouse phospholipid hydroperoxide glutathione peroxidase 4.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6VLY
 
 | | Crystal structure of FabI-NADH complex from Alistipes finegoldii | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Radka, C.D, Seetharaman, J, Rock, C.O. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The genome of a Bacteroidetes inhabitant of the human gut encodes a structurally distinct enoyl-acyl carrier protein reductase (FabI).
J.Biol.Chem., 295, 2020
|
|
8RCC
 
 | | W-formate dehydrogenase from Desulfovibrio vulgaris - aerobic soaked with 48 bar CO2 for 1 min | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CARBON DIOXIDE, ... | | Authors: | Vilela-Alves, G, Carpentier, P, Manuel, R.R, Pereira, I.C, Romao, M.J, Mota, C. | | Deposit date: | 2023-12-06 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Substrate-dependent oxidative inactivation of a W-dependent formate dehydrogenase involving selenocysteine displacement.
Chem Sci, 15, 2024
|
|
7QUL
 
 | | Alcohol Dehydrogenase from Thauera aromatica K319A/K320A mutant | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, ZINC ION | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7ZZG
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},24~{R},25~{S},26~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-25,26-bis(oxidanyl)-27-oxa-2,4,6,9,14,17,20,22-octazatetracyclo[22.2.1.0^{2,10}.0^{3,8}]heptacosa-3(8),4,6,9-tetraen-11-yne-16,21-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZF
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-26-oxa-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3(8),4,6,9-tetraen-11-yne-18,20-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZE
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},25~{R},26~{S},27~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-26,27-bis(oxidanyl)-28-oxa-2,4,6,9,14,17,21,23-octazatetracyclo[23.2.1.0^{2,10}.0^{3,8}]octacosa-3(8),4,6,9-tetraen-11-yne-16,22-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZH
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},22~{R},23~{S},24~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-23,24-bis(oxidanyl)-25-oxa-2,4,6,9,14,17,20-heptazatetracyclo[20.2.1.0^{2,10}.0^{3,8}]pentacosa-3(8),4,6,9-tetraen-11-yne-16,19-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
7ZZJ
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a linear di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-26-oxa-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3(8),4,6,9-tetraen-11-yne-16,20-dione, CITRIC ACID, NAD kinase 1 | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
9DRS
 
 | | Crystal structure of M. tuberculosis PheRS-tRNA complex bound to inhibitor D-116 | | Descriptor: | 1,2-ETHANEDIOL, 1H-benzimidazol-2-amine, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Gade, P, Chang, C, Forte, B, Wower, J, Gilbert, I.H, Baragana, B, Michalska, K, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
7QNH
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE (III) ION, GLYCEROL, ... | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R.K, Widersten, M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and kinetic studies of a laboratory evolved aldehyde reductase explain the dramatic shift of its new substrate specificity.
Iucrj, 10, 2023
|
|
9GV8
 
 | |
9DQI
 
 | | D306N Mutant of M.tuberculosis MenD (SEPHCHC Synthase) | | Descriptor: | 1,2-ETHANEDIOL, 1,4-dihydroxy-2-naphthoic acid, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, ... | | Authors: | Johnston, J.M, Ho, N.A.T, Given, F.M, Bulloch, E.M.M, Allison, T.M, Jiao, W. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Apparent Reversal of Allosteric Response in Mycobacterium tuberculosis MenD Reveals Links to Half-of-Sites Reactivity.
Chembiochem, 26, 2025
|
|
6I6F
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 1 | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Sepiapterin reductase, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8WFP
 
 | | Crystal structure of polo-like kinase(PLK1)PBD in complex with DD-1 | | Descriptor: | DD-1, Serine/threonine-protein kinase PLK1 | | Authors: | Park, J, La, Y.K, Bang, J.K, Lee, S.J. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Degradation of Polo-like Kinase 1 by the Novel Poly-Arginine N-Degron Pathway PROTAC Regulates Tumor Growth in Nonsmall Cell Lung Cancer.
J.Med.Chem., 67, 2024
|
|
8A1G
 
 | | Structure of the SNX1-SNX5 complex | | Descriptor: | N-PROPANOL, Sorting nexin-1, Sorting nexin-5 | | Authors: | Lopez-Robles, C, Scaramuzza, S, Astorga-Simon, E.N, Banos-Mateos, S, Vidaurrazaga, A, Rojas, A.L, Castano, D, Hierro, A. | | Deposit date: | 2022-06-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture of the ESCPE-1 membrane coat.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OZ4
 
 | | Populus tremula stable protein 1 with an alternate crystal lattice | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-27 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties.
Small Sci, 4, 2024
|
|
5N4Z
 
 | |
5UGG
 
 | | Protease Inhibitor | | Descriptor: | Nalpha-[trans-4-(aminomethyl)cyclohexane-1-carbonyl]-N-octyl-O-[(quinolin-2-yl)methyl]-L-tyrosinamide, Plasminogen | | Authors: | Law, R.H.P, Wu, G, Whisstock, J.C. | | Deposit date: | 2017-01-08 | | Release date: | 2017-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | X-ray crystal structure of plasmin with tranexamic acid-derived active site inhibitors.
Blood Adv, 1, 2017
|
|
6KVJ
 
 | | Crystal structure of UDPX-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
