6XIN
 
 | | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site. | | Descriptor: | (2R,3Z)-3-amino-3-imino-2-[(E)-phenyldiazenyl]propanamide, 1,2-ETHANEDIOL, CESIUM ION, ... | | Authors: | Hilario, E, Chang, C, Mueller, L.J, Dunn, M.F, Fan, L. | | Deposit date: | 2020-06-20 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of tryptophan synthase from Salmonella enterica serovar typhimurium in complex with (2S)-3-Amino-3-imino-2-phenyldiazenylpropanamide at the enzyme alpha-site.
To Be Published
|
|
7Q0P
 
 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
2JAM
 
 | | Crystal structure of human calmodulin-dependent protein kinase I G | | Descriptor: | 1,2-ETHANEDIOL, 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, CALCIUM ION, ... | | Authors: | Debreczeni, J.E, Bullock, A, Keates, T, Niesen, F.H, Salah, E, Shrestha, L, Smee, C, Sobott, F, Pike, A.C.W, Bunkoczi, G, von Delft, F, Turnbull, A, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2006-11-29 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Calmodulin-Dependent Protein Kinase I G
To be Published
|
|
7AOK
 
 | | Crystal structure of CI2 mutant L49I | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
6ODC
 
 | | Crystal structure of HDAC8 in complex with compound 30 | | Descriptor: | (2E)-3-[2-(3-cyclopentyl-5,5-dimethyl-2-oxoimidazolidin-1-yl)phenyl]-N-hydroxyprop-2-enamide, 1,2-ETHANEDIOL, Histone deacetylase 8, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
1A51
 
 | | LOOP D/LOOP E ARM OF E. COLI 5S RRNA, NMR, 9 STRUCTURES | | Descriptor: | 5S RRNA LOOP D/LOOP E | | Authors: | Dallas, A, Moore, P.B. | | Deposit date: | 1998-02-19 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The loop E-loop D region of Escherichia coli 5S rRNA: the solution structure reveals an unusual loop that may be important for binding ribosomal proteins.
Structure, 5, 1997
|
|
9D7R
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Fva1 antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
9D7S
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Api antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
8PPZ
 
 | | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-[(~{E})-2-(2-chlorophenyl)ethenyl]-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Meyners, C, Deutscher, R.C.E, Hausch, F. | | Deposit date: | 2023-07-10 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Co-crystal structure of FKBP12, compound 7 and the FRB fragment of mTOR
Chemrxiv, 2023
|
|
7A1H
 
 | | Crystal structure of wild-type CI2 | | Descriptor: | SULFATE ION, Subtilisin-chymotrypsin inhibitor-2A | | Authors: | Olsen, J.G, Teilum, K, Hamborg, L, Roche, J.V. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synergistic stabilization of a double mutant in chymotrypsin inhibitor 2 from a library screen in E. coli.
Commun Biol, 4, 2021
|
|
9DFC
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with lasso peptide lariocidin, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Travin, D.Y, Jangra, M, Kaur, M, Darwish, L, Koteva, K, Klepacki, D, Wang, W, Tiffany, M, Sokaribo, A, Coombes, B.K, Vazquez-Laslop, N, Wright, G.D, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome.
Res Sq, 2024
|
|
5LS7
 
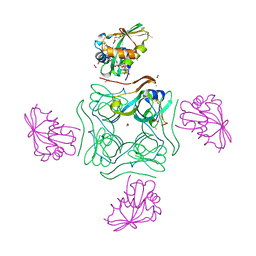 | | Complex of wild type E. coli alpha aspartate decarboxylase with its processing factor PanZ | | Descriptor: | ACETYL COENZYME *A, Aspartate 1-decarboxylase, CARBON DIOXIDE, ... | | Authors: | Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The Mechanism of Regulation of Pantothenate Biosynthesis by the PanD-PanZAcCoA Complex Reveals an Additional Mode of Action for the Antimetabolite N-Pentyl Pantothenamide (N5-Pan).
Biochemistry, 56, 2017
|
|
6YN0
 
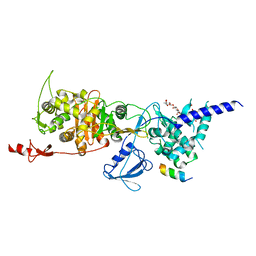 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | Descriptor: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | Deposit date: | 2020-04-10 | | Release date: | 2020-11-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
9D7T
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with Api137 antimicrobial peptide, mRNA, A-site release factor 1, and deacylated P-site and E-site tRNAphe at 2.70A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Huang, W, Baliga, C, Atkinson, G.C, Vazquez-Laslop, N, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-17 | | Release date: | 2024-11-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activity, structure, and diversity of Type II proline-rich antimicrobial peptides from insects.
Embo Rep., 25, 2024
|
|
6OXI
 
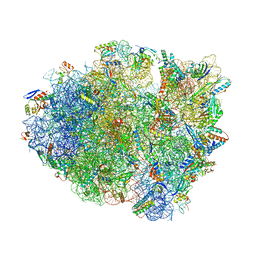 | | Dimeric E.coli YoeB bound to Thermus thermophilus 70S post-cleavage (UAA) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pavelich, I.J, Hoffer, E.D, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.495 Å) | | Cite: | Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability.
Nucleic Acids Res., 47, 2019
|
|
6ND5
 
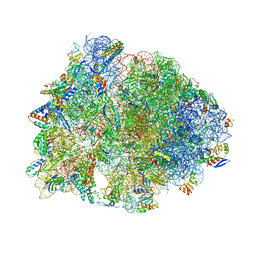 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with chloramphenicol and bound to mRNA and A-, P-, and E-site tRNAs at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-03-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
6ND6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with erythromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.85A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Plessa, E, Chen, C.-W, Bougas, A, Krokidis, M.G, Dinos, G.P, Polikanov, Y.S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-02-20 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | High-resolution crystal structures of ribosome-bound chloramphenicol and erythromycin provide the ultimate basis for their competition.
RNA, 25, 2019
|
|
9C12
 
 | | Leukotriene C4-bound Multidrug Resistance-associated Protein 2 (MRP2) | | Descriptor: | (5~{S},6~{R},7~{E},9~{E},11~{Z},14~{Z})-6-[(2~{R})-2-[[(4~{S})-4-azanyl-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]-3-( 2-hydroxy-2-oxoethylamino)-3-oxidanylidene-propyl]sulfanyl-5-oxidanyl-icosa-7,9,11,14-tetraenoic acid, ATP-binding cassette sub-family C member 2, CHOLESTEROL, ... | | Authors: | Koide, E, Chen, J. | | Deposit date: | 2024-05-28 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Structural basis for the transport and regulation mechanism of the multidrug resistance-associated protein 2.
Nat Commun, 16, 2025
|
|
7PZY
 
 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
6QE5
 
 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
7Q0R
 
 | | Structure of the Candida albicans 80S ribosome in complex with blasticidin s | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
9D0J
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with BT-33, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | (4S,5aS,8S,8aR)-N-[(1R,4R,6Z,8R,9R,10R,11R,12S,13R)-4-fluoro-11,12,13-trihydroxy-8-methyl-14-oxa-2-thiabicyclo[8.3.1]tetradec-6-en-9-yl]-4-(2-methylpropyl)octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Tresco, B.I.C, Wu, K.J.Y, Ramkissoon, A, Purdy, M, See, D.N.Y, Liu, R.Y, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2024-08-07 | | Release date: | 2024-11-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of a fluorinated macrobicyclic antibiotic through chemical synthesis.
Nat.Chem., 17, 2025
|
|
6OTR
 
 | | Dimeric E.coli YoeB bound to Thermus thermophilus 70S post-cleavage (AAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pavelich, I.P, Hoffer, E.D, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-03 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability.
Nucleic Acids Res., 47, 2019
|
|
6OXA
 
 | | Dimeric E.coli YoeB bound to Thermus thermophilus 70S pre-cleavage (AAU) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pavelich, I.J, Hoffer, E.D, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2019-05-13 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Monomeric YoeB toxin retains RNase activity but adopts an obligate dimeric form for thermal stability.
Nucleic Acids Res., 47, 2019
|
|
7A3M
 
 | |
