7BV2
 
 | | The nsp12-nsp7-nsp8 complex bound to the template-primer RNA and triphosphate form of Remdesivir(RTP) | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yin, W, Mao, C, Luan, X, Shen, D, Shen, Q, Su, H, Wang, X, Zhou, F, Zhao, W, Gao, M, Chang, S, Xie, Y.C, Tian, G, Jiang, H.W, Tao, S.C, Shen, J, Jiang, Y, Jiang, H, Xu, Y, Zhang, S, Zhang, Y, Xu, H.E. | | Deposit date: | 2020-04-09 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir.
Science, 368, 2020
|
|
2XI5
 
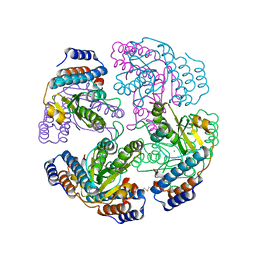 | | N-terminal endonuclease domain of La Crosse virus L-protein | | Descriptor: | MANGANESE (II) ION, RNA POLYMERASE L | | Authors: | Reguera, J, Weber, F, Cusack, S. | | Deposit date: | 2010-06-28 | | Release date: | 2010-10-06 | | Last modified: | 2012-07-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bunyaviridae RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral CAP-Dependent Transcription.
Plos Pathog., 6, 2010
|
|
2XI7
 
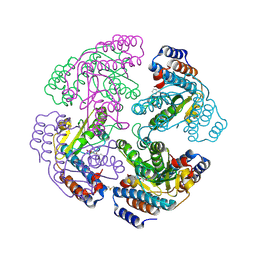 | | N-terminal endonuclease domain of La Crosse virus L-protein | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, RNA POLYMERASE L | | Authors: | Reguera, J, Weber, F, Cusack, S. | | Deposit date: | 2010-06-28 | | Release date: | 2010-10-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bunyaviridae RNA Polymerases (L-Protein) Have an N-Terminal, Influenza-Like Endonuclease Domain, Essential for Viral CAP-Dependent Transcription.
Plos Pathog., 6, 2010
|
|
8PM0
 
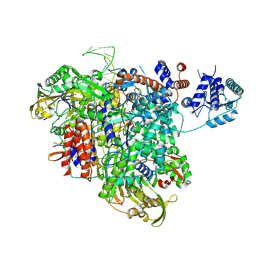 | |
8R3L
 
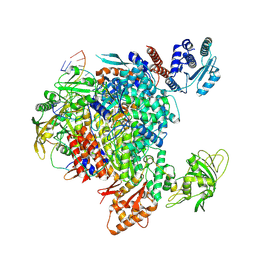 | | Influenza A/H7N9 polymerase in pre-initiation state, intermediate conformation (I) with PB2-C(I), ENDO(T), and Pol II pS5 CTD peptide mimic bound in site 1A/2A | | Descriptor: | 3' vRNA end (51-mer vRNA loop), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-11-09 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The host RNA polymerase II C-terminal domain is the anchor for replication of the influenza virus genome.
Nat Commun, 15, 2024
|
|
8R3K
 
 | |
3UGP
 
 | |
1TQL
 
 | | POLIOVIRUS POLYMERASE G1A MUTANT | | Descriptor: | ACETIC ACID, RNA-directed RNA polymerase | | Authors: | Thompson, A.A, Peersen, O.B. | | Deposit date: | 2004-06-17 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for proteolysis-dependent activation of the poliovirus RNA-dependent RNA polymerase
Embo J., 23, 2004
|
|
3UGO
 
 | |
3C2P
 
 | |
3C3L
 
 | |
3C46
 
 | |
1P16
 
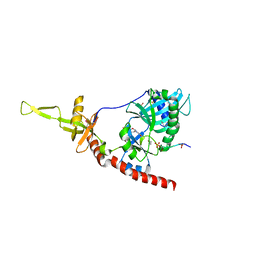 | | Structure of an mRNA capping enzyme bound to the phosphorylated carboxyl-terminal domain of RNA polymerase II | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Fabrega, C, Shen, V, Shuman, S, Lima, C.D. | | Deposit date: | 2003-04-11 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II.
Mol.Cell, 11, 2003
|
|
4GZK
 
 | |
1HMJ
 
 | | SOLUTION STRUCTURE OF RNA POLYMERASE SUBUNIT H | | Descriptor: | PROTEIN (SUBUNIT H) | | Authors: | Thiru, A, Hodach, M, Eloranta, J, Kostourou, V, Weinzierl, R. | | Deposit date: | 1999-02-05 | | Release date: | 1999-04-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA polymerase subunit H features a beta-ribbon motif within a novel fold that is present in archaea and eukaryotes.
J.Mol.Biol., 287, 1999
|
|
4FF3
 
 | | N4 mini-vRNAP transcription initiation complex, 3 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
4FF4
 
 | |
4FF1
 
 | |
4FF2
 
 | | N4 mini-vRNAP transcription initiation complex, 2 min after soaking GTP, ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bacteriophage N4 P2 promoter, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Murakami, K.S, Basu, R.S. | | Deposit date: | 2012-05-30 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Watching the Bacteriophage N4 RNA Polymerase Transcription by Time-dependent Soak-trigger-freeze X-ray Crystallography.
J.Biol.Chem., 288, 2013
|
|
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | Descriptor: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | Authors: | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | Deposit date: | 2020-09-04 | | Release date: | 2020-09-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3E3J
 
 | | Crystal Structure of an Intermediate Complex of T7 RNAP and 8nt of RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*DTP*DAP*DAP*DTP*DAP*DCP*DGP*DAP*DCP*DTP*DCP*DAP*DCP*DTP*DAP*DTP*DAP*DTP*DTP*DTP*DCP*DTP*DGP*DCP*DCP*DAP*DAP*DAP*DCP*DGP*DGP*DC)-3'), DNA-directed RNA polymerase, ... | | Authors: | Durniak, K.J, Bailey, S, Steitz, T.A. | | Deposit date: | 2008-08-07 | | Release date: | 2008-11-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Science, 322, 2008
|
|
4NJC
 
 | |
7NFR
 
 | | Fujian capmidlink domain in complex with Nb8194 | | Descriptor: | GLYCEROL, NB8194, Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NFQ
 
 | | Fujian capmidlink domain in complex with Nb8193 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Nb8193, ... | | Authors: | Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2021-02-07 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NFT
 
 | |
