7S96
 
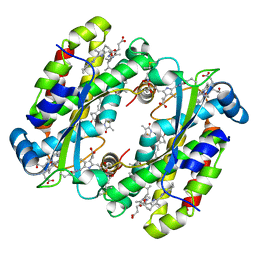 | | Structure of the Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
1MOE
 
 | | The three-dimensional structure of an engineered scFv T84.66 dimer or diabody in VL to VH linkage. | | Descriptor: | SULFATE ION, anti-CEA mAb T84.66 | | Authors: | Carmichael, J.A, Power, B.E, Garrett, T.P.J, Yazaki, P.J, Shively, J.E, Raubischek, A.A, Wu, A.M, Hudson, P.J. | | Deposit date: | 2002-09-09 | | Release date: | 2003-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of an Anti-CEA scFv Diabody Assembled from T84.66 scFvs in VL-to-VH Orientation: Implications for Diabody Flexibility
J.Mol.Biol., 326, 2003
|
|
9GUD
 
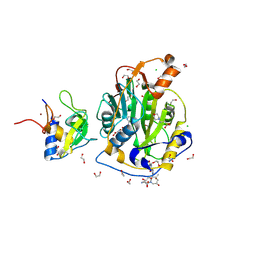 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54570922 | | Descriptor: | (3~{S})-3-azanyl-4-[(3~{R},4~{R},6~{S})-3-[1,3-dimethyl-2,6-bis(oxidanylidene)purin-7-yl]-4-methyl-4,6-bis(oxidanyl)azepan-1-yl]-4-oxidanylidene-butanoic acid, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
1MP4
 
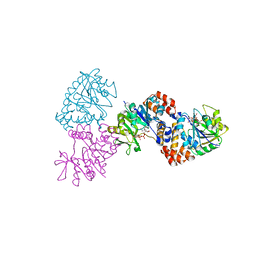 | | W224H VARIANT OF S. ENTERICA RmlA | | Descriptor: | URIDINE-5'-DIPHOSPHATE-GLUCOSE, W224H Variant of S. Enterica RmlA Bound to UDP-Glucose | | Authors: | Barton, W.A, Biggins, J.B, Jiang, J, Thorson, J.S, Nikolov, D.B. | | Deposit date: | 2002-09-11 | | Release date: | 2002-10-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Expanding pyrimidine diphosphosugar libraries via structure-based nucleotidylyltransferase engineering
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
9GUY
 
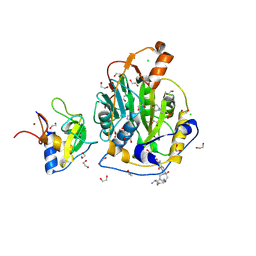 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571098 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-20 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
1MPE
 
 | |
9H86
 
 | | Small circular RNA dimer - Class 4 | | Descriptor: | Circular RNA, Complementary strand | | Authors: | McRae, E.K, Kristoffersen, E.L, Holliger, P, Andersen, E.S. | | Deposit date: | 2024-10-28 | | Release date: | 2024-12-04 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Roles of dimeric intermediates in RNA-catalyzed rolling circle synthesis.
Nucleic Acids Res., 53, 2025
|
|
1M02
 
 | | NMR Structure of PW2 Bound to SDS Micelles: A Tryptophan-rich Anticocidial Peptide Selected from Phage Display Libraries | | Descriptor: | HIS-PRO-LEU-LYS-GLN-TYR-TRP-TRP-ARG-PRO-SER-ILE | | Authors: | Tinoco, L.W, da Silva Jr, A, Leite, A, Valente, A.P, Almeida, F.C. | | Deposit date: | 2002-06-11 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of PW2 bound to SDS micelles. A tryptophan-rich anticoccidial peptide selected from phage display libraries
J.Biol.Chem., 277, 2002
|
|
5FF1
 
 | | Two way mode of binding of antithyroid drug methimazole to mammalian heme peroxidases: Structure of the complex of lactoperoxidase with methimazole at 1.97 Angstrom resolution | | Descriptor: | 1-METHYL-1,3-DIHYDRO-2H-IMIDAZOLE-2-THIONE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, R.P, Singh, A, Sirohi, H, Singh, A.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Dual binding mode of antithyroid drug methimazole to mammalian heme peroxidases - structural determination of the lactoperoxidase-methimazole complex at 1.97 angstrom resolution.
Febs Open Bio, 6, 2016
|
|
1M0V
 
 | | NMR STRUCTURE OF THE TYPE III SECRETORY DOMAIN OF YERSINIA YOPH COMPLEXED WITH THE SKAP-HOM PHOSPHO-PEPTIDE N-acetyl-DEpYDDPF-NH2 | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, SKAP55 homologue | | Authors: | Khandelwal, P, Keliikuli, K, Smith, C.L, Saper, M.A, Zuiderweg, E.R.P. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-24 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure and phosphopeptide binding to the N-terminal domain of Yersinia YopH: comparison with a crystal structure
Biochemistry, 41, 2002
|
|
9GUF
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571106 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9GUE
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 57256189 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
9GWD
 
 | | ZT-KP6-1: AN EFFECTOR FROM ZYMOSEPTORIA TRITICI | | Descriptor: | Zt-KP6-1 | | Authors: | Barthe, P, de Guillen, K. | | Deposit date: | 2024-09-26 | | Release date: | 2024-10-09 | | Last modified: | 2025-09-17 | | Method: | SOLUTION NMR | | Cite: | Zymoseptoria tritici Effectors Structurally Related to Killer Proteins UmV-KP4 and UmV-KP6 Inhibit Fungal Growth, and Define Extended Protein Families in Fungi.
Mol Plant Pathol, 26, 2025
|
|
1MLN
 
 | |
9GUT
 
 | | 30S mRNA delivery complex (bS1 resolved) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
1MMD
 
 | | TRUNCATED HEAD OF MYOSIN FROM DICTYOSTELIUM DISCOIDEUM COMPLEXED WITH MGADP-BEF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Fisher, A.J, Holden, H.M, Rayment, I. | | Deposit date: | 1995-03-21 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4-.
Biochemistry, 34, 1995
|
|
9GUP
 
 | | 30S mRNA delivery complex (open head) | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
1MB6
 
 | |
9H36
 
 | |
1M2V
 
 | |
9H35
 
 | |
1M3I
 
 | | Perfringolysin O, new crystal form | | Descriptor: | perfringolysin O | | Authors: | Rossjohn, J, Parker, M, Polekhina, G, Feil, S, Tweten, R. | | Deposit date: | 2002-06-28 | | Release date: | 2004-02-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Snapshots in the Molecular Mechanism of PFO Revealed
To be Published
|
|
1M46
 
 | |
9GUX
 
 | | 30S-TEC (TEC in expressome position) Inactive state 1 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
