6IU3
 
 | | Crystal structure of iron transporter VIT1 with zinc ions | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Taniguchi, R, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
7K5V
 
 | | OXA-48 bound by Compound 3.1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Taylor, D.M, Hu, L, Prasad, B.V.V, Palzkill, T. | | Deposit date: | 2020-09-17 | | Release date: | 2021-12-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Unique Diacidic Fragments Inhibit the OXA-48 Carbapenemase and Enhance the Killing of Escherichia coli Producing OXA-48.
Acs Infect Dis., 7, 2021
|
|
3VWX
 
 | | Structural analysis of an epsilon-class glutathione S-transferase from housefly, Musca domestica | | Descriptor: | GLUTATHIONE, Glutathione s-transferase 6B | | Authors: | Nakamura, C, Sue, M, Miyamoto, T, Yajima, S. | | Deposit date: | 2012-09-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of an epsilon-class glutathione transferase from housefly, Musca domestica
Biochem.Biophys.Res.Commun., 430, 2013
|
|
6IU6
 
 | | Crystal structure of cytoplasmic metal binding domain with nickel ions | | Descriptor: | NICKEL (II) ION, VIT1, ZINC ION | | Authors: | Kato, T, Nishizawa, T, Yamashita, K, Kumazaki, K, Ishitani, R, Nureki, O. | | Deposit date: | 2018-11-27 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of plant vacuolar iron transporter VIT1.
Nat Plants, 5, 2019
|
|
3WWO
 
 | | S-selective hydroxynitrile lyase from Baliospermum montanum (apo1) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (S)-hydroxynitrile lyase, CALCIUM ION | | Authors: | Nakano, S, Dadashipour, M, Asano, Y. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and functional analysis of hydroxynitrile lyase from Baliospermum montanum with crystal structure, molecular dynamics and enzyme kinetics
Biochim.Biophys.Acta, 1844, 2014
|
|
2J9H
 
 | | Crystal structure of human glutathione-S-transferase P1-1 cys-free mutant in complex with S-hexylglutathione at 2.4 A resolution | | Descriptor: | GLUTATHIONE S-TRANSFERASE P, S-HEXYLGLUTATHIONE | | Authors: | Tars, K, Hegazy, U.M, Hellman, U, Mannervik, B. | | Deposit date: | 2006-11-08 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Modulating Catalytic Activity by Unnatural Amino Acid Residues in a Gsh-Binding Loop of Gst P1-1.
J.Mol.Biol., 376, 2008
|
|
2J9Y
 
 | | Tryptophan Synthase Q114N mutant in complex with Compound II | | Descriptor: | (3E)-4-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}-2-IMINOBUT-3-ENOIC ACID, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Blumenstein, L, Domratcheva, T, Niks, D, Ngo, H, Seidel, R, Dunn, M.F, Schlichting, I. | | Deposit date: | 2006-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Betaq114N and Betat110V Mutations Reveal a Critically Important Role of the Substrate Alpha-Carboxylate Site in the Reaction Specificity of Tryptophan Synthase.
Biochemistry, 46, 2007
|
|
2J9Z
 
 | | Tryptophan Synthase T110 mutant complex | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Blumenstein, L, Domratcheva, T, Niks, D, Ngo, H, Seidel, R, Dunn, M.F, Schlichting, I. | | Deposit date: | 2006-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Betaq114N and Betat110V Mutations Reveal a Critically Important Role of the Substrate Alpha-Carboxylate Site in the Reaction Specificity of Tryptophan Synthase.
Biochemistry, 46, 2007
|
|
4XYD
 
 | | Nitric oxide reductase from Roseobacter denitrificans (RdNOR) | | Descriptor: | CALCIUM ION, COPPER (II) ION, FE (III) ION, ... | | Authors: | Crow, A. | | Deposit date: | 2015-02-02 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the Membrane-intrinsic Nitric Oxide Reductase from Roseobacter denitrificans.
Biochemistry, 55, 2016
|
|
4YCL
 
 | | Crystal structure of the SR CA2+-ATPASE with bound CPA | | Descriptor: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ogawa, H, Takahashi, M, Kondou, Y, Toyoshima, C. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4DZ5
 
 | |
1UA3
 
 | |
1WBJ
 
 | | wildtype tryptophan synthase complexed with glycerol phosphate | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SN-GLYCEROL-3-PHOSPHATE, SODIUM ION, ... | | Authors: | Kulik, V, Weyand, M, Schlichting, I. | | Deposit date: | 2004-11-01 | | Release date: | 2006-05-24 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | On the Structural Basis of the Catalytic Mechanism and the Regulation of the Alpha Subunit of Tryptophan Synthase from Salmonella Typhimurium and Bx1 from Maize, Two Evolutionarily Related Enzymes.
J.Mol.Biol., 352, 2005
|
|
4DJ9
 
 | | Human vinculin head domain Vh1 (residues 1-258) in complex with the talin vinculin binding site 50 (VBS50, residues 2078-2099) | | Descriptor: | Talin-1, Vinculin | | Authors: | Yogesha, S.D, Rangarajan, E.S, Vonrhein, C, Bricogne, G, Izard, T. | | Deposit date: | 2012-02-01 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of vinculin in complex with vinculin binding site 50 (VBS50), the integrin binding site 2 (IBS2) of talin.
Protein Sci., 21, 2012
|
|
4DBS
 
 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 (AKR1C3) in complex with NADP+ and 3'-[(4-nitronaphthalen-1-yl)amino]benzoic acid | | Descriptor: | 3-[(4-nitronaphthalen-1-yl)amino]benzoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, M, Christianson, D.W, Winkler, J.D, Penning, T.M. | | Deposit date: | 2012-01-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Crystal structures of AKR1C3 containing an N-(aryl)amino-benzoate inhibitor and a bifunctional AKR1C3 inhibitor and androgen receptor antagonist. Therapeutic leads for castrate resistant prostate cancer.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3ZM0
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
1PBE
 
 | |
1VF2
 
 | |
1VYO
 
 | |
3ZM2
 
 | | Catalytic domain of human SHP2 | | Descriptor: | TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 11 | | Authors: | Bohm, K, Schuetz, A, Roske, Y, Heinemann, U. | | Deposit date: | 2013-02-04 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Selective Inhibitors of the Protein Tyrosine Phosphatase Shp2 Block Cellular Motility and Growth of Cancer Cells in Vitro and in Vivo.
Chemmedchem, 10, 2015
|
|
1VZK
 
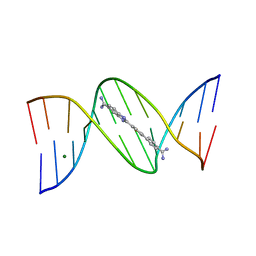 | | A Thiophene Based Diamidine Forms a "Super" AT Binding Minor Groove Agent | | Descriptor: | 2-(5-{4-[AMINO(IMINO)METHYL]PHENYL}-2-THIENYL)-1H-BENZIMIDAZOLE-6- CARBOXIMIDAMIDE DIHYDROCHLORIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP *CP*G)-3', MAGNESIUM ION | | Authors: | Mallena, S, Lee, M.P.H, Bailly, C, Neidle, S, Kumar, A, Boykin, D.W, Wilson, W.D. | | Deposit date: | 2004-05-20 | | Release date: | 2004-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thiophene-Based Diamidine Forms a "Super" at Binding Minor Groove Agent
J.Am.Chem.Soc., 142, 2004
|
|
7L2G
 
 | |
6HBV
 
 | |
1RWL
 
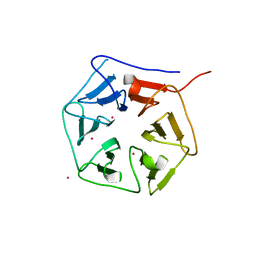 | | Extracellular domain of Mycobacterium tuberculosis PknD | | Descriptor: | CADMIUM ION, Serine/threonine-protein kinase pknD | | Authors: | Good, M.C, Greenstein, A.E, Young, T.A, Ng, H.L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-12-16 | | Release date: | 2004-04-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sensor Domain of the Mycobacterium tuberculosis Receptor Ser/Thr Protein Kinase, PknD, forms a Highly Symmetric beta Propeller.
J.Mol.Biol., 339, 2004
|
|
2JGS
 
 | | Circular permutant of avidin | | Descriptor: | BIOTIN, CIRCULAR PERMUTANT OF AVIDIN | | Authors: | Maatta, J.A.E, Hytonen, V.P, Airenne, T.T, Niskanen, E, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-02-14 | | Release date: | 2008-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Modification of Ligand-Binding Preference of Avidin by Circular Permutation and Mutagenesis.
Chembiochem, 9, 2008
|
|
