5T2Q
 
 | |
4Z1L
 
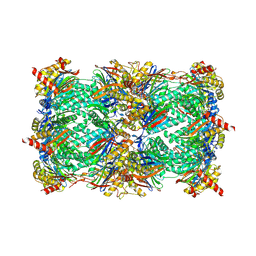 | | Yeast 20S proteasome in complex with belactosin C derivative 3 | | Descriptor: | (2S,3S)-2-{(1R)-2-[(3,5-dimethoxybenzyl)amino]-1-hydroxy-2-oxoethyl}-3-methylpentanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Minimal beta-Lactone Fragment for Selective beta 5c or beta 5i Proteasome Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4G7U
 
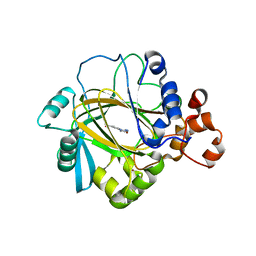
 | | Rat Heme Oxygenase-1 in complex with Heme and CO with 16 hr Illumination: Laser off | | Descriptor: | CARBON MONOXIDE, FORMIC ACID, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Moffat, K, Noguchi, M. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discrimination between CO and O(2) in heme oxygenase: comparison of static structures and dynamic conformation changes following CO photolysis.
Biochemistry, 51, 2012
|
|
3BG6
 
 | | Pyranose 2-oxidase from Trametes multicolor, E542K mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design
Febs J., 276, 2009
|
|
2XNS
 
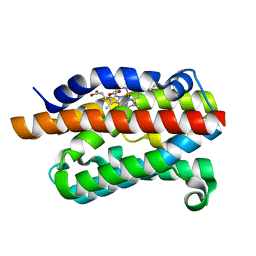
 | | Crystal Structure Of Human G alpha i1 Bound To A Designed Helical Peptide Derived From The Goloco Motif Of RGS14 | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G(I) SUBUNIT ALPHA-1, GUANOSINE-5'-DIPHOSPHATE, REGULATOR OF G-PROTEIN SIGNALING 14, ... | | Authors: | Bosch, D, Sammond, D.W, Butterfoss, G.L, Machius, M, Siderovski, D.P, Kuhlman, B. | | Deposit date: | 2010-08-05 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Computational Design of the Sequence and Structure of a Protein-Binding Peptide.
J.Am.Chem.Soc., 133, 2011
|
|
4GD4
 
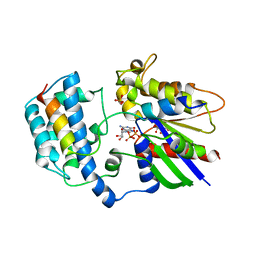
 | | Crystal Structure of JMJD2A Complexed with Inhibitor | | Descriptor: | 2-(1H-pyrazol-3-yl)pyridine-4-carboxylic acid, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | King, O.N.F, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, McDonough, M.A, Schofield, C.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-07-31 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of JMJD2A Complexed with Inhibitor
TO BE PUBLISHED
|
|
5SY6
 
 | | Atomic resolution structure of human DJ-1, DTT bound | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Protein deglycase DJ-1 | | Authors: | Wilson, M.A, Lin, J. | | Deposit date: | 2016-08-10 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Short Carboxylic Acid-Carboxylate Hydrogen Bonds Can Have Fully Localized Protons.
Biochemistry, 56, 2017
|
|
5T2R
 
 | |
5T4O
 
 | | Autoinhibited E. coli ATP synthase state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
4Z9O
 
 | | Crystal Structure of human GGT1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Terzyan, S.S, Hanigan, M.H. | | Deposit date: | 2015-04-10 | | Release date: | 2015-06-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human gamma-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.
J.Biol.Chem., 290, 2015
|
|
5AEU
 
 | | Crystal structure of II9 variant of Biphenyl dioxygenase from Burkholderia xenovorans LB400 | | Descriptor: | BIPHENYL DIOXYGENASE SUBUNIT ALPHA, BIPHENYL DIOXYGENASE SUBUNIT BETA, FE (II) ION, ... | | Authors: | Dhindwal, S, Gomez-Gil, L, Sylvestre, M, Eltis, L.D, Bolin, J.T, Kumar, P. | | Deposit date: | 2015-01-10 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of the Enhanced Pollutant-Degrading Capabilities of an Engineered Biphenyl Dioxygenase
J.Bacteriol., 198, 2016
|
|
2XND
 
 | | Crystal structure of bovine F1-c8 sub-complex of ATP Synthase | | Descriptor: | ATP SYNTHASE LIPID-BINDING PROTEIN, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT ALPHA, ... | | Authors: | Watt, I.N, Montgomery, M.G, Runswick, M.J, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2010-08-02 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Bioenergetic Cost of Making an Adenosine Triphosphate Molecule in Animal Mitochondria.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5ARI
 
 | | Bovine mitochondrial ATP synthase state 2b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
7DDI
 
 | | Crystal structures of Na+,K+-ATPase in complex with digitoxin | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ogawa, H, Cornelius, F, Kanai, R, Motoyama, K, Vilsen, B, Toyoshima, C. | | Deposit date: | 2020-10-29 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.72 Å) | | Cite: | Binding of cardiotonic steroids to Na + ,K + -ATPase in the E2P state.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2ZPU
 
 | | Crystal Structure of Modified Serine Racemase from S.pombe. | | Descriptor: | MAGNESIUM ION, N-(5'-PHOSPHOPYRIDOXYL)-D-ALANINE, Uncharacterized protein C320.14 | | Authors: | Goto, M. | | Deposit date: | 2008-07-29 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Serine racemase with catalytically active lysinoalanyl residue.
J.Biochem., 145, 2009
|
|
5E0S
 
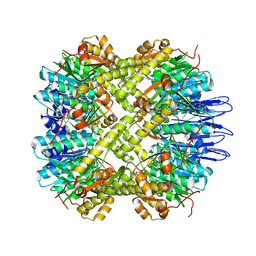 | | crystal structure of the active form of the proteolytic complex clpP1 and clpP2 | | Descriptor: | ATP-dependent Clp protease proteolytic subunit 1, ATP-dependent Clp protease proteolytic subunit 2 | | Authors: | LI, M, Wlodawer, A, Maurizi, M. | | Deposit date: | 2015-09-29 | | Release date: | 2016-02-17 | | Last modified: | 2016-04-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Functional Properties of the Active Form of the Proteolytic Complex, ClpP1P2, from Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5TBE
 
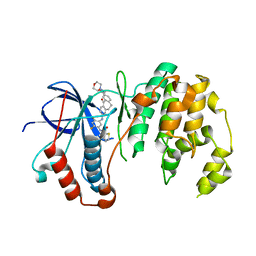 | | Human p38alpha MAP Kinase in Complex with Dibenzosuberone Compound 2 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[2,4-bis(fluoranyl)-5-[[9-(2-morpholin-4-ylethylcarbamoyl)-11-oxidanylidene-5,6-dihydrodibenzo[1,2-~{d}:1',2'-~{f}][7]annulen-3-yl]amino]phenyl]thiophene-2-carboxamide | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-09-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimized Target Residence Time: Type I1/2 Inhibitors for p38 alpha MAP Kinase with Improved Binding Kinetics through Direct Interaction with the R-Spine.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ARE
 
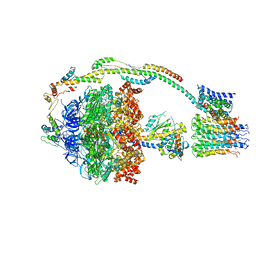 | | Bovine mitochondrial ATP synthase state 1b | | Descriptor: | ATP SYNTHASE F(0) COMPLEX SUBUNIT B1, MITOCHONDRIAL, ATP SYNTHASE F(0) COMPLEX SUBUNIT C1, ... | | Authors: | Zhou, A, Rohou, A, Schep, D.G, Bason, J.V, Montgomery, M.G, Walker, J.E, Grigorieff, N, Rubinstein, J.L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM.
Elife, 4, 2015
|
|
4ZTH
 
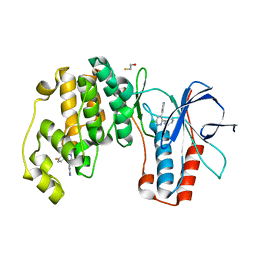 | | Structure of human p38aMAPK-arylpyridazinylpyridine fragment complex used in inhibitor discovery | | Descriptor: | 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, 6-chloro-3-phenyl-4-(pyridin-4-yl)pyridazine, BETA-MERCAPTOETHANOL, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Watterson, D.M. | | Deposit date: | 2015-05-14 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Selective and Brain Penetrant p38 alpha MAPK Inhibitor Candidate for Neurologic and Neuropsychiatric Disorders That Attenuates Neuroinflammation and Cognitive Dysfunction.
J.Med.Chem., 62, 2019
|
|
5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
3AON
 
 | | Crystal structure of the central axis (NtpD-NtpG) in the catalytic portion of Enterococcus hirae V-type sodium ATPase | | Descriptor: | NITRATE ION, V-type sodium ATPase subunit D, V-type sodium ATPase subunit G | | Authors: | Saijo, S, Arai, S, Hossain, K.M.M, Yamato, I, Kakinuma, Y, Ishizuka-Katsura, Y, Terada, T, Shirouzu, M, Yokoyama, S, Iwata, S, Murata, T. | | Deposit date: | 2010-10-04 | | Release date: | 2011-10-05 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the central axis DF complex of the prokaryotic V-ATPase
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5VG1
 
 | |
1A6B
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN THE ZINC FINGER PROTEIN NCP10 OF MOLONEY MURINE LEUKEMIA VIRUS AND A SEQUENCE OF THE PSI-PACKAGING DOMAIN OF HIV-1, 20 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*CP*GP*CP*C)-3'), ZINC FINGER PROTEIN NCP10, ZINC ION | | Authors: | Schueler, W, Dong, C.-Z, Wecker, K, Roques, B.P. | | Deposit date: | 1998-02-23 | | Release date: | 1999-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the complex between the zinc finger protein NCp10 of Moloney murine leukemia virus and the single-stranded pentanucleotide d(ACGCC): comparison with HIV-NCp7 complexes.
Biochemistry, 38, 1999
|
|
1AB7
 
 | |
3DQA
 
 | |
