8U4R
 
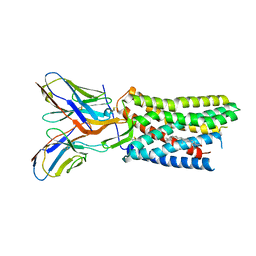 | |
8U4P
 
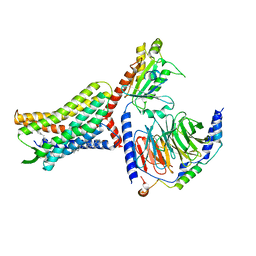 | | Structure of AMD3100-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4O
 
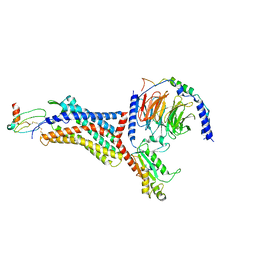 | | Structure of CXCL12-bound CXCR4/Gi complex | | Descriptor: | C-X-C chemokine receptor type 4, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Saotome, K, McGoldrick, L.L, Franklin, M.C. | | Deposit date: | 2023-09-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural insights into CXCR4 modulation and oligomerization.
Nat.Struct.Mol.Biol., 2024
|
|
8U4S
 
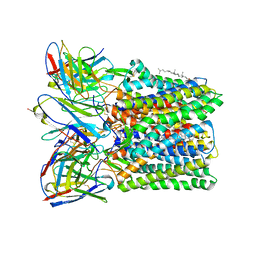 | |
8U4Q
 
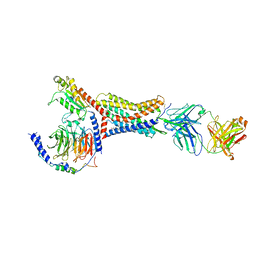 | |
8T6L
 
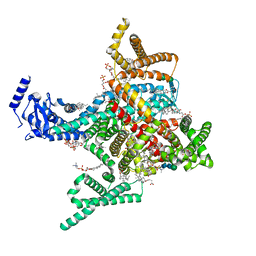 | | Cryo-EM structure of rat cardiac sodium channel NaV1.5 with batrachotoxin analog BTX-B | | Descriptor: | (1R)-1-[(5aR,7aR,9R,11aS,11bS,12R,13aR)-9,12-dihydroxy-2,11a-dimethyl-1,2,3,4,7a,8,9,10,11,11a,12,13-dodecahydro-7H-9,11b-epoxy-13a,5a-prop[1]enophenanthro[2,1-f][1,4]oxazepin-14-yl]ethyl benzoate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, ... | | Authors: | Tonggu, L, Wisedchaisri, G, Gamal El-Din, T.M, Zheng, N, Catterall, W.A. | | Deposit date: | 2023-06-16 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dual receptor-sites reveal the structural basis for hyperactivation of sodium channels by poison-dart toxin batrachotoxin.
Nat Commun, 15, 2024
|
|
8XLD
 
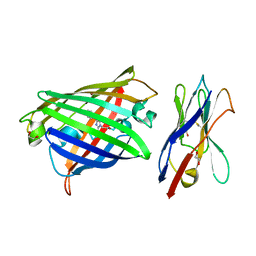 | | Structure of the GFP:GFP-nanobody complex from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Nanobody(Staygold-S2G10)-Nanobody(Staygold-S4F1), ZINC ION, ... | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Bao, C. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the GFP:GFP-nanobody complex from Biortus.
To Be Published
|
|
8SXC
 
 | |
8TU9
 
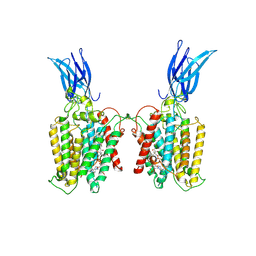 | | Cryo-EM structure of HGSNAT-acetyl-CoA complex at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL COENZYME *A, Enhanced green fluorescent protein,Heparan-alpha-glucosaminide N-acetyltransferase,Isoform 2 of Heparan-alpha-glucosaminide N-acetyltransferase | | Authors: | Navratna, V, Kumar, A, Mosalaganti, S. | | Deposit date: | 2023-08-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the human heparan-alpha-glucosaminide N -acetyltransferase (HGSNAT).
Elife, 13, 2024
|
|
8C7I
 
 | |
8U23
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 8 | | Descriptor: | Green Fluorescent Protein Variant #8, ccGFP 8 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U22
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 7 | | Descriptor: | Green Fluorescent Protein Variant #7, ccGFP 7 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U20
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 5 | | Descriptor: | Green Fluorescent Protein Variant #5, ccGFP 5 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U21
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP E6 | | Descriptor: | Green Fluorescent Protein Variant E6, ccGFP E6 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8U24
 
 | | A Highly Stable Variant of Corynactis Californica Green Fluorescent Protein, ccGFP 9 | | Descriptor: | Green Fluorescent Protein Variant #9, ccGFP 9 | | Authors: | Hung, L.-W, Terwilliger, T.C, Waldo, G, Nguyen, H.B. | | Deposit date: | 2023-09-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering highly stable variants of Corynactis californica green fluorescent proteins.
Protein Sci., 33, 2024
|
|
8DHR
 
 | | An ester mutant of SfGFP | | Descriptor: | Green fluorescent protein | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
8C1X
 
 | |
8OVY
 
 | | Structure of analogue of superfolded GFP | | Descriptor: | Green fluorescent protein | | Authors: | Dunkelmann, D, Fiedler, M, Bellini, D, Alvira, C.P, Chin, J.W. | | Deposit date: | 2023-04-26 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | Adding alpha , alpha-disubstituted and beta-linked monomers to the genetic code of an organism.
Nature, 625, 2024
|
|
8QBJ
 
 | | Structure of mBaoJin at pH 4.6 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8QDD
 
 | | Structure of mBaoJin at pH 8.5 | | Descriptor: | CHLORIDE ION, SULFATE ION, mBaoJin | | Authors: | Samygina, V.R, Vlaskina, A.V, Gabdulkhakov, A, Subach, O.M, Subach, F.V. | | Deposit date: | 2023-08-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8Q79
 
 | | Structure of mBaoJin at pH 6.5 | | Descriptor: | CHLORIDE ION, mBaoJin | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Nikolaeva, A.Y, Borshchevsky, V, Qin, W, Subach, F.V. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bright and stable monomeric green fluorescent protein derived from StayGold.
Nat.Methods, 21, 2024
|
|
8BXP
 
 | |
8TLM
 
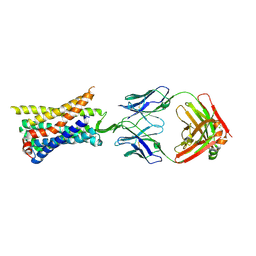 | | Structure of a class A GPCR/Fab complex | | Descriptor: | C-C chemokine receptor type 8, Green fluorescent protein fusion, Fab heavy chain, ... | | Authors: | Sun, D, Johnson, M, Masureel, M. | | Deposit date: | 2023-07-27 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of antibody inhibition and chemokine activation of the human CC chemokine receptor 8.
Nat Commun, 14, 2023
|
|
8J2K
 
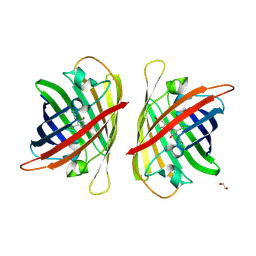 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutation (N137A, Q140S) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(N137A, Q140S) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
8J2J
 
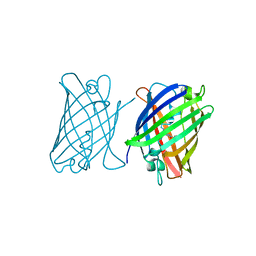 | | Crystal structure of a bright green fluorescent protein (StayGold) with single mutation (Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, StayGold(Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
