7AFZ
 
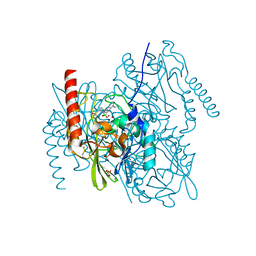 | | L1 metallo-b-lactamase with compound EBL-1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
7AFX
 
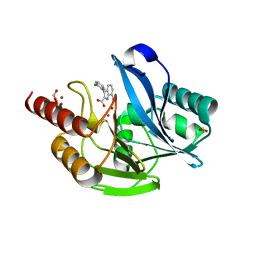 | |
7AEZ
 
 | |
7AFY
 
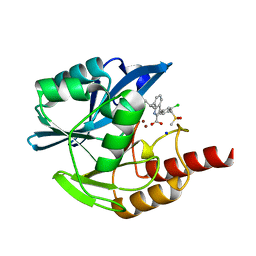 | | Crystal structure of the metallo-beta-lactamase VIM1 with 1306 | | Descriptor: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, SODIUM ION, ... | | Authors: | Brem, J, Schofield, C.J. | | Deposit date: | 2020-09-21 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.105 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM2 with 139
To Be Published
|
|
7LUU
 
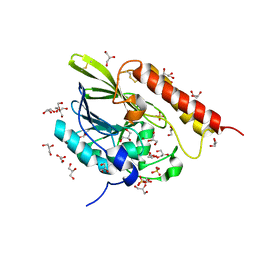 | | Kinetic and Structural Characterization of the First B3 Metallo-beta-Lactamase with an Active Site Glutamic Acid | | Descriptor: | GLYCEROL, SULFATE ION, Subclass B3 metallo-beta-lactamase, ... | | Authors: | Wilson, L, Knaven, E, Morris, M.T, Schenk, G. | | Deposit date: | 2021-02-23 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Kinetic and Structural Characterization of the First B3 Metallo-beta-Lactamase with an Active-Site Glutamic Acid.
Antimicrob.Agents Chemother., 65, 2021
|
|
7L91
 
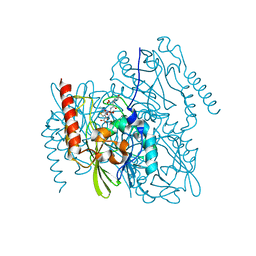 | | Structure of Metallo Beta-Lactamase L1 in a Complex with Hydrolyzed Moxalactam Determined by Pink-Beam Serial Crystallography | | Descriptor: | (2R)-2-[(R)-carboxy{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}methoxymethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-oxazine-4-carboxylic acid, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Wilamowski, M, Kim, Y, Sherrell, D.A, Lavens, A, Henning, R, Maltseva, N, Endres, M, Babnigg, G, Vukica, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-01-01 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Time-resolved beta-lactam cleavage by L1 metallo-beta-lactamase.
Nat Commun, 13, 2022
|
|
4KEQ
 
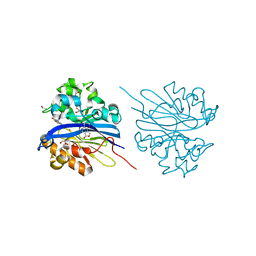 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
8D2Z
 
 | |
3DHC
 
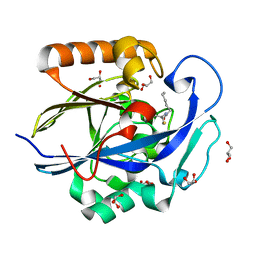 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
4J5F
 
 | | Crystal Structure of B. thuringiensis AiiA mutant F107W | | Descriptor: | GLYCEROL, N-acyl homoserine lactonase, ZINC ION | | Authors: | Liu, C.F, Liu, D, Momb, J, Thomas, P.W, Lajoie, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2013-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-gamma-lactonase from Bacillus thuringiensis.
Biochemistry, 52, 2013
|
|
3DHA
 
 | | An Ultral High Resolution Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at An Alternative Site | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
3DH8
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | FE (III) ION, Uncharacterized protein PA1000, bis(4-nitrophenyl) hydrogen phosphate | | Authors: | Yu, S, Blankenfeldt, W. | | Deposit date: | 2008-06-17 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
3DHB
 
 | | 1.4 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homoserine Bound at The Catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homoserine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
4J5H
 
 | | Crystal Structure of B. thuringiensis AiiA mutant F107W with N-decanoyl-L-homoserine bound at the active site | | Descriptor: | GLYCEROL, N-acyl homoserine lactonase, N-decanoyl-L-homoserine, ... | | Authors: | Liu, C.F, Liu, D, Momb, J, Thomas, P.W, Lajoie, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2013-02-08 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | A phenylalanine clamp controls substrate specificity in the quorum-quenching metallo-gamma-lactonase from Bacillus thuringiensis.
Biochemistry, 52, 2013
|
|
7KGX
 
 | | Structure of PQS Response Protein PqsE in Complex with 4-(3-(2-methyl-2-morpholinobutyl)ureido)-N-(thiazol-2-yl)benzamide | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, 4-({[(2R)-2-methyl-2-(morpholin-4-yl)butyl]carbamoyl}amino)-N-(1,3-thiazol-2-yl)benzamide, FE (III) ION | | Authors: | Jeffrey, P.D, Taylor, I.R, Bassler, B.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitor Mimetic Mutations in the Pseudomonas aeruginosa PqsE Enzyme Reveal a Protein-Protein Interaction with the Quorum-Sensing Receptor RhlR That Is Vital for Virulence Factor Production.
Acs Chem.Biol., 16, 2021
|
|
7KGW
 
 | |
3ESH
 
 | | Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314 | | Descriptor: | Protein similar to metal-dependent hydrolase | | Authors: | Seetharaman, J, Chen, Y, Wang, H, Janjua, H, Foote, E.L, Xiao, R, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-06 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a probable metal-dependent hydrolase from
Staphylococcus aureus. Northeast Structural Genomics target ZR314
To be Published
|
|
4LE6
 
 | | Crystal structure of the phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Organophosphorus hydrolase, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-06-25 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Enzymatic Characterization of the Phosphotriesterase OPHC2 from Pseudomonas pseudoalcaligenes.
Plos One, 8, 2013
|
|
4KEP
 
 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
7O0O
 
 | | Crystal structure of the B3 metallo-beta-lactamase L1 with hydrolysed ertapenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallography and QM/MM Simulations Identify Preferential Binding of Hydrolyzed Carbapenem and Penem Antibiotics to the L1 Metallo-beta-Lactamase in the Imine Form.
J.Chem.Inf.Model., 61, 2021
|
|
7OVH
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 14 (JMV-6931) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVE
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 10 (JMV-7210) | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, UNKNOWN LIGAND, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Mangani, S, Docquier, J.D, Pozzi, C, Marcoccia, F. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
7OVF
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in Complex with compound 8 (JMV-7207) | | Descriptor: | 4-[2-[(4-fluorophenyl)methylsulfanyl]ethyl]-3-phenyl-1H-1,2,4-triazole-5-thione, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Sannio, F, Marcoccia, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-06-14 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1,2,4-Triazole-3-thione compounds with a 4-ethyl alkyl/aryl sulfide substituent are broad-spectrum metallo-beta-lactamase inhibitors with re-sensitization activity.
Eur.J.Med.Chem., 226, 2021
|
|
3Q6V
 
 | | Crystal Structure of Serratia fonticola Sfh-I: glycerol complex | | Descriptor: | Beta-lactamase, GLYCEROL, ZINC ION | | Authors: | Fonseca, F, Saavedra, M.J, Correia, A, Spencer, J. | | Deposit date: | 2011-01-03 | | Release date: | 2011-07-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of Serratia fonticola Sfh-I: activation of the nucleophile in mono-zinc metallo-beta-lactamases.
J.Mol.Biol., 411, 2011
|
|
7PP0
 
 | | Crystal structure of the VIM-2 acquired metallo-beta-Lactamase in complex with compound 28 (JMV-7038) | | Descriptor: | 2-[2-[3-[3-(2-morpholin-4-ylethoxy)phenyl]-5-sulfanylidene-1H-1,2,4-triazol-4-yl]ethyl]benzoic acid, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Tassone, G, Benvenuti, M, Verdirosa, F, Corsica, G, Chelini, G, De Luca, F, Docquier, J.D, Pozzi, C, Mangani, S. | | Deposit date: | 2021-09-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1,2,4-Triazole-3-Thione Analogues with a 2-Ethylbenzoic Acid at Position 4 as VIM-type Metallo-beta-Lactamase Inhibitors.
Chemmedchem, 17, 2022
|
|
