2RQB
 
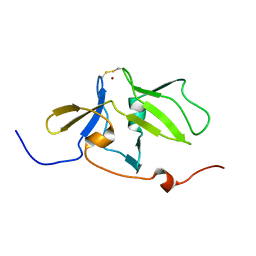 | | Solution structure of MDA5 CTD | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, ZINC ION | | Authors: | Takahasi, K, Kumeta, H, Tsuduki, N, Narita, R, Shigemoto, T, Hirai, R, Yoneyama, M, Horiuchi, M, Ogura, K, Fujita, T, Fuyuhiko, I. | | Deposit date: | 2009-03-17 | | Release date: | 2009-05-05 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Cytosolic RNA Sensor MDA5 and LGP2 C-terminal Domains: IDENTIFICATION OF THE RNA RECOGNITION LOOP IN RIG-I-LIKE RECEPTORS
J.Biol.Chem., 284, 2009
|
|
2RMJ
 
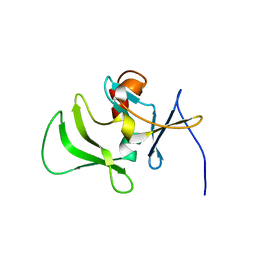 | | Solution structure of RIG-I C-terminal domain | | Descriptor: | Probable ATP-dependent RNA helicase DDX58 | | Authors: | Takahasi, K, Yoneyama, M, Nihishori, T, Hirai, R, Narita, R, Gale Jr, M, Fujita, T, Inagaki, F. | | Deposit date: | 2007-10-23 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nonself RNA-Sensing Mechanism of RIG-I Helicase and Activation of Antiviral Immune Responses
Mol.Cell, 29, 2008
|
|
1UZ5
 
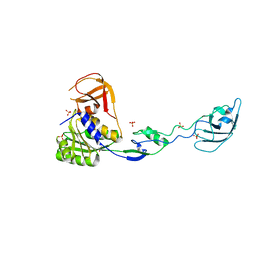 | |
2RQE
 
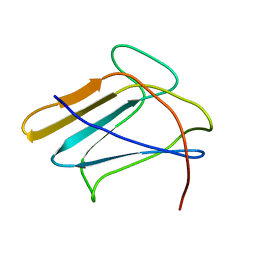 | | Solution structure of the silkworm bGRP/GNBP3 N-terminal domain reveals the mechanism for b-1,3-glucan specific recognition | | Descriptor: | Beta-1,3-glucan-binding protein | | Authors: | Takahasi, K, Ochiai, M, Horiuchi, M, Kumeta, H, Ogura, K, Ashida, M, Inagaki, F. | | Deposit date: | 2009-04-22 | | Release date: | 2009-06-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2YSP
 
 | | Solution structure of the C2H2 type zinc finger (region 507-539) of human Zinc finger protein 224 | | Descriptor: | ZINC ION, Zinc finger protein 224 | | Authors: | Takahashi, M, Kuwasako, K, Tsuda, K, Tanabe, W, Harada, T, Watanabe, S, Tochio, N, Muto, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 507-539)of human Zinc finger protein 224
To be Published
|
|
3A77
 
 | | The crystal structure of phosphorylated IRF-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Interferon regulatory factor 3 | | Authors: | Takahasi, K, Horiuchi, M, Noda, N.N, Inagaki, F. | | Deposit date: | 2009-09-17 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ser386 phosphorylation of transcription factor IRF-3 induces dimerization and association with CBP/p300 without overall conformational change.
Genes Cells, 15, 2010
|
|
4YB4
 
 | | Crystal structure of homoisocitrate dehydrogenase from Thermus thermophilus in complex with homoisocitrate, magnesium ion (II) and NADH | | Descriptor: | (1R,2S)-1-hydroxybutane-1,2,4-tricarboxylic acid, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Takahashi, K, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2015-02-18 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of homoisocitrate dehydrogenase from Thermus thermophilus in complex with homoisocitrate, magnesium(II) and NADH
To Be Published
|
|
5XIT
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form II | | Descriptor: | E3 ubiquitin-protein ligase RNF168, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
5XIS
 
 | | Crystal structure of RNF168 UDM1 in complex with Lys63-linked diubiquitin, form I | | Descriptor: | E3 ubiquitin-protein ligase RNF168, MAGNESIUM ION, Ubiquitin-40S ribosomal protein S27a, ... | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
1GF8
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GAZ
 
 | | Crystal Structure of Mutant Human Lysozyme Substituted at the Surface Positions | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of amino acid residues at turns in the conformational stability and folding of human lysozyme.
Biochemistry, 39, 2000
|
|
1GFE
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GF9
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFJ
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1INU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
5XIU
 
 | | Crystal structure of RNF168 UDM2 in complex with Lys63-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase RNF168, Ubiquitin-40S ribosomal protein S27a | | Authors: | Takahashi, T.S, Sato, Y, Fukai, S. | | Deposit date: | 2017-04-27 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into two distinct binding modules for Lys63-linked polyubiquitin chains in RNF168.
Nat Commun, 9, 2018
|
|
1GFH
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFT
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFV
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFA
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFK
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFU
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFG
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
1GFR
 
 | | CRYSTAL STRUCTURE OF MUTANT HUMAN LYSOZYME SUBSTITUTED AT THE SURFACE POSITIONS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Funahashi, J, Takano, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-12-04 | | Release date: | 2000-12-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Positive contribution of hydration structure on the surface of human lysozyme to the conformational stability.
J.Biol.Chem., 277, 2002
|
|
2E5B
 
 | |
