7ZA8
 
 | | BRD4 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Isoform C of Bromodomain-containing protein 4 | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-03-22 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
1Y83
 
 | |
5GGE
 
 | | Fatty Acid-Binding Protein in Brain Tissue of Drosophila melanogaster | | Descriptor: | CITRIC ACID, Fatty acid bindin protein, isoform B | | Authors: | Cheng, Y.-Y, Huang, Y.-F, Lin, H.-H, Chang, W.W, Lyu, P.-C. | | Deposit date: | 2016-06-15 | | Release date: | 2017-06-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | The ligand-mediated affinity of brain-type fatty acid-binding protein for membranes determines the directionality of lipophilic cargo transport.
Biochim Biophys Acta Mol Cell Biol Lipids, 1864, 2019
|
|
7VWF
 
 | |
7VWG
 
 | |
7VWH
 
 | |
7VWE
 
 | |
5SY7
 
 | | Crystal Structure of the Heterodimeric NPAS3-ARNT Complex with HRE DNA | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, DNA (5'-D(*CP*AP*CP*GP*AP*CP*CP*CP*GP*CP*AP*CP*GP*TP*AP*CP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*GP*CP*TP*GP*CP*GP*TP*AP*CP*GP*TP*GP*CP*GP*GP*GP*TP*CP*GP*T)-3'), ... | | Authors: | Wu, D, Su, X, Potluri, N, Kim, Y, Rastinejad, F. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | NPAS1-ARNT and NPAS3-ARNT crystal structures implicate the bHLH-PAS family as multi-ligand binding transcription factors.
Elife, 5, 2016
|
|
2A0T
 
 | | NMR structure of the FHA1 domain of Rad53 in complex with a biological relevant phosphopeptide derived from Madt1 | | Descriptor: | Hypothetical 73.8 kDa protein in SAS3-SEC17 intergenic region, residues 301-310, Serine/threonine-protein kinase RAD53 | | Authors: | Mahajan, A, Yuan, C, Pike, B.L, Heierhorst, J, Chang, C.-F, Tsai, M.-D. | | Deposit date: | 2005-06-16 | | Release date: | 2005-11-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | FHA Domain-Ligand Interactions: Importance of Integrating Chemical and Biological Approaches
J.Am.Chem.Soc., 127, 2005
|
|
2W87
 
 | | Xyl-CBM35 in complex with glucuronic acid containing disaccharide. | | Descriptor: | CALCIUM ION, ESTERASE D, UNKNOWN LIGAND, ... | | Authors: | Montainer, C, Bueren, A.L.v, Dumon, C, Flint, J.E, Correia, M.A, Prates, J.A, Firbank, S.J, Lewis, R.J, Grondin, G.G, Ghinet, M.G, Gloster, T.M, Herve, C, Knox, J.P, Talbot, B.G, Turkenburg, J.P, Kerovuo, J, Brzezinski, R, Fontes, C.M.G.A, Davies, G.J, Boraston, A.B, Gilbert, H.J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-01-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evidence that Family 35 Carbohydrate Binding Modules Display Conserved Specificity But Divergent Function.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8B2C
 
 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by an epoxide derivative | | Descriptor: | (1~{S},2~{R},4~{R},5~{S},6~{S})-2,4,5-trihydroxy-7-oxabicyclo[4.1.0]heptane-2-carboxylic acid, 3-dehydroquinate dehydratase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2A
 
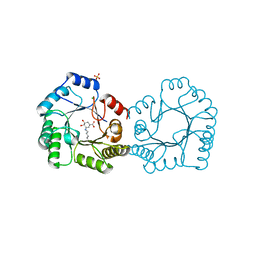 | | Crystal structure of type I dehydroquinase from Staphylococcus aureus inhibited by a hydroxylamine derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, CHLORIDE ION, ... | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
8B2B
 
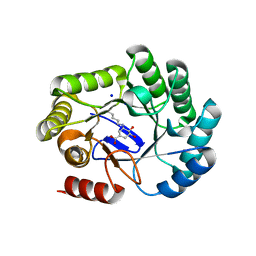 | | Crystal structure of type I dehydroquinase from Salmonella typhi inhibited by a hydroxylamine derivative | | Descriptor: | (4R,5R)-3-amino-4,5-dihydroxy-cyclohexene-1-carboxylic acid, 3-dehydroquinate dehydratase, SODIUM ION | | Authors: | Otero, J.M, Rodriguez, A, Maneiro, M, Lence, E, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2022-09-13 | | Release date: | 2023-02-15 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quinate-based ligands for irreversible inactivation of the bacterial virulence factor DHQ1 enzyme-A molecular insight.
Front Mol Biosci, 10, 2023
|
|
1QL9
 
 | |
5O8V
 
 | |
4U0C
 
 | |
4U0B
 
 | |
4U0A
 
 | | Hexameric HIV-1 CA in complex with CPSF6 peptide, P6 crystal form | | Descriptor: | CHLORIDE ION, Capsid protein p24, Cleavage and polyadenylation specificity factor subunit 6 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
4U0D
 
 | | Hexameric HIV-1 CA in complex with Nup153 peptide, P212121 crystal form | | Descriptor: | CHLORIDE ION, Gag polyprotein, Nuclear pore complex protein Nup153 | | Authors: | Price, A.J, Jacques, D.A, James, L.C. | | Deposit date: | 2014-07-11 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host Cofactors and Pharmacologic Ligands Share an Essential Interface in HIV-1 Capsid That Is Lost upon Disassembly.
Plos Pathog., 10, 2014
|
|
5D2M
 
 | | Complex between human SUMO2-RANGAP1, UBC9 and ZNF451 | | Descriptor: | 1,2-ETHANEDIOL, Ran GTPase-activating protein 1, SUMO-conjugating enzyme UBC9, ... | | Authors: | Cappadocia, L, Lima, C.D. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for catalytic activation by the human ZNF451 SUMO E3 ligase.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3HJO
 
 | |
3HJM
 
 | | Crystal structure of human Glutathione Transferase Pi Y108V mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
3RS1
 
 | | Mouse C-type lectin-related protein Clrg | | Descriptor: | C-type lectin domain family 2 member I, CHLORIDE ION | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Stepankova, A, Koval, T, Hasek, J, Kotynkova, K, Vanek, O, Bezouska, K. | | Deposit date: | 2011-05-02 | | Release date: | 2012-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mouse Clr-g, a Ligand for NK Cell Activation Receptor NKR-P1F: Crystal Structure and Biophysical Properties.
J.Immunol., 189, 2012
|
|
3R1L
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Mg2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', Class I ligase ribozyme, MAGNESIUM ION, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.125 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1F4B
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI THYMIDYLATE SYNTHASE | | Descriptor: | GLYCEROL, SULFATE ION, THYMIDYLATE SYNTHASE | | Authors: | Erlanson, D.A, Braisted, A.C, Raphael, D.R, Randal, M, Stroud, R.M, Gordon, E, Wells, J.A. | | Deposit date: | 2000-06-07 | | Release date: | 2000-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Site-directed ligand discovery.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
