2K9P
 
 | |
2KEG
 
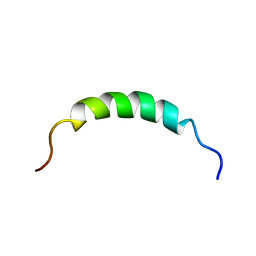 | | NMR structure of Plantaricin K in DPC-micelles | | Descriptor: | PlnK | | Authors: | Rogne, P, Haugen, M, Fimland, G, Nissen-Meyer, J, Kristiansen, P. | | Deposit date: | 2009-01-30 | | Release date: | 2009-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the two-peptide bacteriocin plantaricin JK.
Peptides, 30, 2009
|
|
2N3B
 
 | | Structure of oxidized horse heart cytochrome c encapsulated in reverse micelles | | Descriptor: | Cytochrome c, HEME C | | Authors: | O'Brien, E.S, Nucci, N.V, Fuglestad, B, Tommos, C, Wand, A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-10-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN.
J.Biol.Chem., 290, 2015
|
|
2KNE
 
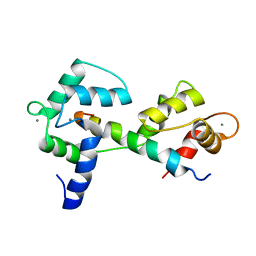 | | Calmodulin wraps around its binding domain in the plasma membrane CA2+ pump anchored by a novel 18-1 motif | | Descriptor: | ATPase, Ca++ transporting, plasma membrane 4, ... | | Authors: | Juranic, N, Atanasova, E, Filoteo, A.G, Macura, S, Prendergast, F.G, Penniston, J.T, Strehler, E.E. | | Deposit date: | 2009-08-21 | | Release date: | 2009-11-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Calmodulin wraps around its binding domain in the plasma membrane Ca2+ pump anchored by a novel 18-1 motif.
J.Biol.Chem., 285, 2010
|
|
2KHG
 
 | |
2L2B
 
 | |
2O9P
 
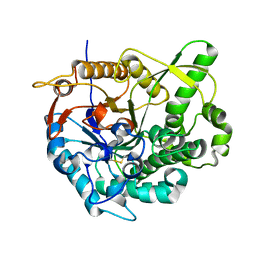 | | beta-glucosidase B from Paenibacillus polymyxa | | Descriptor: | Beta-glucosidase B | | Authors: | Isorna, P, Polaina, J, Sanz-Aparicio, J. | | Deposit date: | 2006-12-14 | | Release date: | 2007-10-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Paenibacillus polymyxa beta-Glucosidase B Complexes Reveal the Molecular Basis of Substrate Specificity and Give New Insights into the Catalytic Machinery of Family I Glycosidases
J.Mol.Biol., 371, 2007
|
|
7UQZ
 
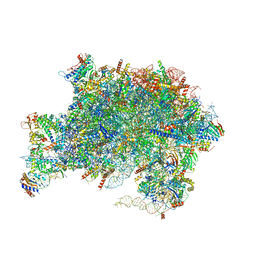 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1 D52A strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
8EWX
 
 | |
4WK8
 
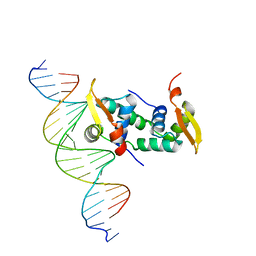 | | FOXP3 forms a domain-swapped dimer to bridge DNA | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3'), DNA (5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3'), Forkhead box protein P3 | | Authors: | Chen, Y, Chen, L. | | Deposit date: | 2014-10-01 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions.
Nucleic Acids Res., 43, 2015
|
|
4PU2
 
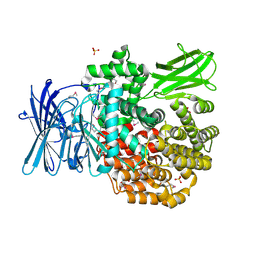 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine L-(R)-LeuP | | Descriptor: | Aminopeptidase N, GLYCEROL, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine
To be Published
|
|
4WMR
 
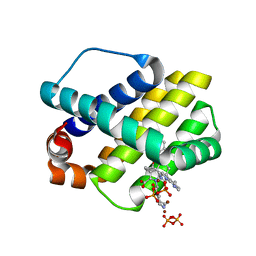 | | STRUCTURE OF MCL1 BOUND TO BRD inhibitor ligand 1 AT 1.7A | | Descriptor: | 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, PYROPHOSPHATE 2-, ... | | Authors: | CLIFTON, M.C, EDWARDS, T.E. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WMX
 
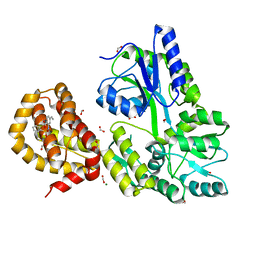 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
7UQB
 
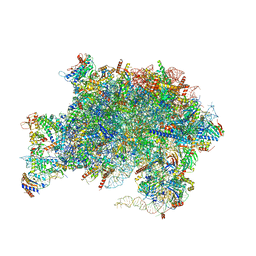 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1-D52A strain with AlF4 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
4PLP
 
 | |
8AYE
 
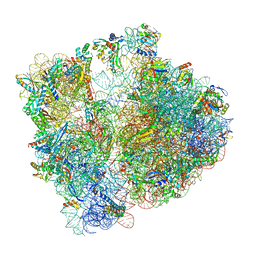 | | E. coli 70S ribosome bound to thermorubin and fMet-tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Sanyal, S, Parajuli, N.P, Emmerich, A.G. | | Deposit date: | 2022-09-02 | | Release date: | 2023-03-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | Antibiotic thermorubin tethers ribosomal subunits and impedes A-site interactions to perturb protein synthesis in bacteria.
Nat Commun, 14, 2023
|
|
4PL8
 
 | |
6LYZ
 
 | |
8AVF
 
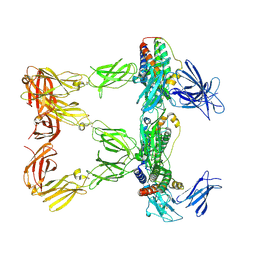 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (6.45 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVO
 
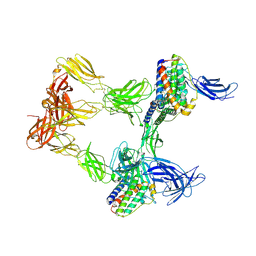 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4PVB
 
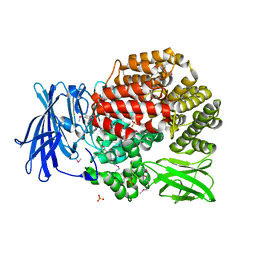 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP) | | Descriptor: | Aminopeptidase N, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-16 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine (D-(S)-LeuP)
To be Published
|
|
8B6H
 
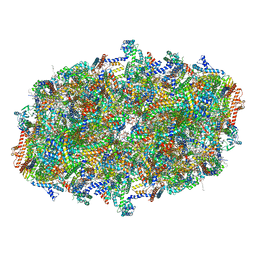 | | Cryo-EM structure of cytochrome c oxidase dimer (complex IV) from respiratory supercomplex of Tetrahymena thermophila | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2022-09-27 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-12 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of mitochondrial membrane bending by the I-II-III 2 -IV 2 supercomplex.
Nature, 615, 2023
|
|
8AVC
 
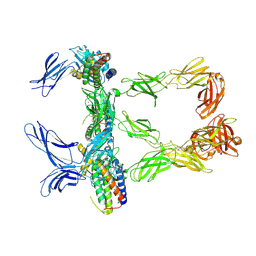 | | Mouse leptin:LEP-R complex cryoEM structure (3:3 model) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVD
 
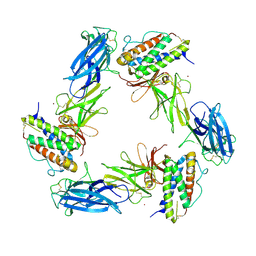 | | Cryo-EM structure for a 3:3 complex between mouse leptin and the mouse LEP-R ectodomain (local refinement) | | Descriptor: | Leptin, Leptin receptor, NICKEL (II) ION | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4PJ6
 
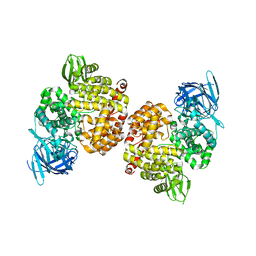 | | Crystal Structure of Human Insulin Regulated Aminopeptidase with Lysine in Active Site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LYSINE, ... | | Authors: | Hermans, S.J, Ascher, D.B, Hancock, N.C, Holien, J.K, Michell, B, Morton, C.J, Parker, M.W. | | Deposit date: | 2014-05-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal structure of human insulin-regulated aminopeptidase with specificity for cyclic peptides.
Protein Sci., 24, 2015
|
|
