6SL3
 
 | |
6SL2
 
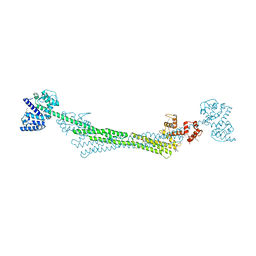 | | ALPHA-ACTININ FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Calponin homology domain protein putative, ... | | Authors: | Pinotsis, N, Khan, M.B, Djinovic-Carugo, K. | | Deposit date: | 2019-08-18 | | Release date: | 2020-08-26 | | Last modified: | 2020-11-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Calcium modulates the domain flexibility and function of an alpha-actinin similar to the ancestral alpha-actinin.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2VYP
 
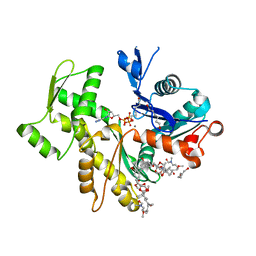 | | Rabbit-muscle G-actin in complex with myxobacterial rhizopodin | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hagelueken, G, Albrecht, S.C, Steinmetz, H, Jansen, R, Heinz, D.W, Kalesse, M, Schubert, W.-D. | | Deposit date: | 2008-07-25 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Absolute Configuration of Rhizopodin and its Inhibition of Actin Polymerization by Dimerization.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
2P9L
 
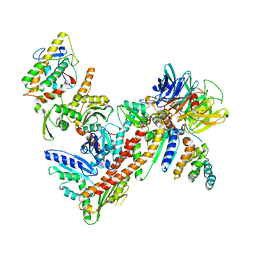 | | Crystal Structure of bovine Arp2/3 complex | | Descriptor: | Actin-like protein 2, Actin-like protein 3, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Nolen, B.J, Pollard, T.D. | | Deposit date: | 2007-03-26 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Insights into the Influence of Nucleotides on Actin Family Proteins from Seven Structures of Arp2/3 Complex.
Mol.Cell, 26, 2007
|
|
2P9U
 
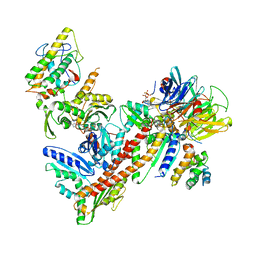 | |
2KRH
 
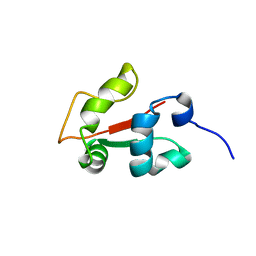 | |
2DJ7
 
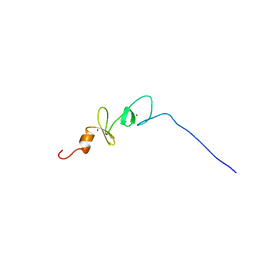 | | Solution Structure of 3rd LIM Domain of Actin-binding LIM Protein 3 | | Descriptor: | Actin-binding LIM protein 3, ZINC ION | | Authors: | Niraula, T.N, Sasagawa, A, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-31 | | Release date: | 2006-10-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of 3rd LIM Domain of Actin-binding LIM Protein 3
To be Published
|
|
5AEY
 
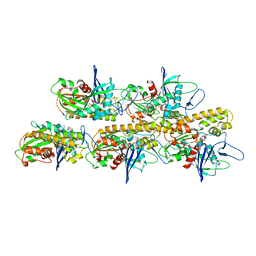 | | actin-like ParM protein bound to AMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PLASMID SEGREGATION PROTEIN PARM | | Authors: | Bharat, T.A.M, Murshudov, G.N, Sachse, C, Lowe, J. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of Actin-Like Parm Filaments Show Architecture of Plasmid-Segregating Spindles.
Nature, 523, 2015
|
|
1V6G
 
 | | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2 | | Descriptor: | Actin Binding LIM Protein 2, ZINC ION | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-11-29 | | Release date: | 2004-06-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the LIM Domain of the Human Actin Binding LIM Protein 2
To be Published
|
|
2CT4
 
 | | Solution Strutcure of the SH3 domain of the Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Miyamoto, K, Tomizawa, T, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-23 | | Release date: | 2005-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Strutcure of the SH3 domain of the Cdc42-interacting protein 4
To be Published
|
|
1WLX
 
 | |
6NW2
 
 | | Structure of human RIPK1 kinase domain in complex with compound 11 | | Descriptor: | (5R)-5-methyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-4,5,6,7-tetrahydro-2H-indazole-3-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Fong, R, Lupardus, P.J. | | Deposit date: | 2019-02-05 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent and selective inhibitors of receptor-interacting protein kinase 1 that lack an aromatic back pocket group.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6GR0
 
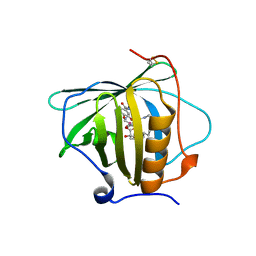 | | Petrobactin-binding engineered lipocalin in complex with gallium-petrobactin | | Descriptor: | 4-[4-[3-[[3,4-bis(oxidanyl)phenyl]carbonylamino]propylamino]butylamino]-2-[2-[4-[3-[[3,4-bis(oxidanyl)phenyl]carbonylamino]propylamino]butylamino]-2-oxidanylidene-ethyl]-2-oxidanyl-4-oxidanylidene-butanoic acid, GALLIUM (III) ION, Neutrophil gelatinase-associated lipocalin | | Authors: | Skerra, A, Eichinger, A. | | Deposit date: | 2018-06-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Reprogramming Human Siderocalin To Neutralize Petrobactin, the Essential Iron Scavenger of Anthrax Bacillus.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7XMK
 
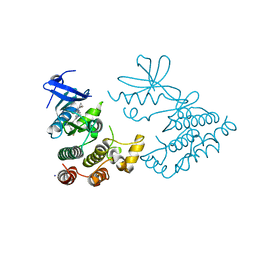 | | Crystal structure of human RIPK1 kinase domain in complex with compound SKLB923 | | Descriptor: | 5-[2-(cyclopropylcarbonylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-(3-fluorophenyl)ethyl]-1-methyl-indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Yang, S. | | Deposit date: | 2022-04-26 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | From Hit to Lead: Structure-Based Optimization of Novel Selective Inhibitors of Receptor-Interacting Protein Kinase 1 (RIPK1) for the Treatment of Inflammatory Diseases.
J.Med.Chem., 67, 2024
|
|
1X64
 
 | |
5DZR
 
 | |
3M6R
 
 | |
5E52
 
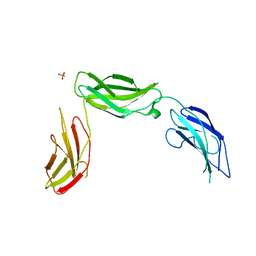 | | Crystal structure of human CNTN5 FN1-FN3 domains | | Descriptor: | Contactin-5, PHOSPHATE ION | | Authors: | Nikolaienko, R.M, Bouyain, S. | | Deposit date: | 2015-10-07 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Structural Basis for Interactions Between Contactin Family Members and Protein-tyrosine Phosphatase Receptor Type G in Neural Tissues.
J.Biol.Chem., 291, 2016
|
|
5E7L
 
 | |
3M6Q
 
 | |
8FIU
 
 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
4G8T
 
 | | Crystal structure of a glucarate dehydratase related protein, from actinobacillus succinogenes, target EFI-502312, with sodium and sulfate bound, ordered loop | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a glucarate dehydratase related protein, from actinobacillus succinogenes, target efi-502312, with sodium and sulfate bound, ordered loop
TO BE PUBLISHED
|
|
1X7G
 
 | | Actinorhodin Polyketide Ketoreductase, act KR, with NADP bound | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
1X7H
 
 | | Actinorhodin Polyketide Ketoreductase, with NADPH bound | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative ketoacyl reductase | | Authors: | Korman, T.P, Hill, J.A, Vu, T.N. | | Deposit date: | 2004-08-13 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of actinorhodin polyketide ketoreductase: cofactor binding and substrate specificity
Biochemistry, 43, 2004
|
|
3V7Z
 
 | | Carboxypeptidase T with GEMSA | | Descriptor: | (2-GUANIDINOETHYLMERCAPTO)SUCCINIC ACID, CALCIUM ION, Carboxypeptidase T, ... | | Authors: | Kuznetsov, S.A, Timofeev, V.I, Akparov, V.K, Kuranova, I.P. | | Deposit date: | 2011-12-22 | | Release date: | 2012-12-26 | | Last modified: | 2015-09-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the broad substrate specificity of carboxypeptidase T from Thermoactinomyces vulgaris.
Febs J., 282, 2015
|
|
