8K00
 
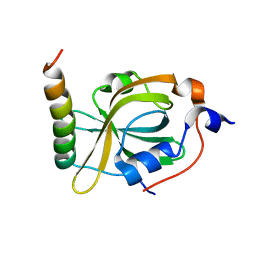 | | RPA70N-MRE11 fusion | | Descriptor: | Double-strand break repair protein MRE11, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Fu, W.M, Wu, Y.Y, Zhou, C. | | Deposit date: | 2023-07-07 | | Release date: | 2023-09-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
3FDQ
 
 | |
8V7G
 
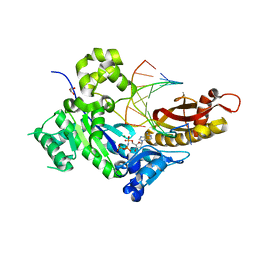 | | Human DNA polymerase eta-DNA-gemC-ended primer-dAMPNPP ternary complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*(U7B))-3'), DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Human DNA polymerase eta-DNA-araC-ended primer-dAMPNPP ternary complex with Mn2+
To Be Published
|
|
1GCB
 
 | | GAL6, YEAST BLEOMYCIN HYDROLASE DNA-BINDING PROTEASE (THIOL) | | Descriptor: | GAL6 HG (EMTS) DERIVATIVE, GLYCEROL, MERCURY (II) ION, ... | | Authors: | Joshua-Tor, L, Xu, H.E, Johnston, S.A, Rees, D.C. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6.
Science, 269, 1995
|
|
7BZH
 
 | |
1BIX
 
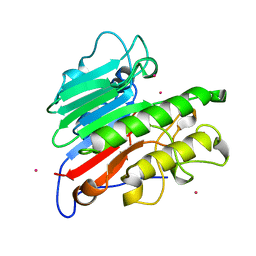 | | THE CRYSTAL STRUCTURE OF THE HUMAN DNA REPAIR ENDONUCLEASE HAP1 SUGGESTS THE RECOGNITION OF EXTRA-HELICAL DEOXYRIBOSE AT DNA ABASIC SITES | | Descriptor: | AP ENDONUCLEASE 1, PLATINUM (II) ION, SAMARIUM (III) ION | | Authors: | Gorman, M.A, Morera, S, Rothwell, D.G, De La Fortelle, E, Mol, C.D, Tainer, J.A, Hickson, I.D, Freemont, P.S. | | Deposit date: | 1998-06-19 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the human DNA repair endonuclease HAP1 suggests the recognition of extra-helical deoxyribose at DNA abasic sites.
EMBO J., 16, 1997
|
|
5UM9
 
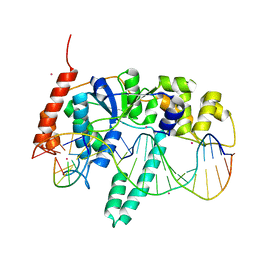 | | Flap endonuclease 1 (FEN1) D86N with 5'-flap substrate DNA and Sm3+ | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*CP*C)-3'), DNA (5'-D(*AP*CP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*T)-3'), DNA (5'-D(P*TP*CP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*GP*T)-3'), ... | | Authors: | Tsutakawa, S.E, Arvai, A.S, Tainer, J.A. | | Deposit date: | 2017-01-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Phosphate steering by Flap Endonuclease 1 promotes 5'-flap specificity and incision to prevent genome instability.
Nat Commun, 8, 2017
|
|
4NDH
 
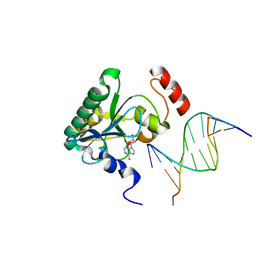 | | Human Aprataxin (Aptx) bound to DNA, AMP, and Zn - product complex | | Descriptor: | 5'-D(P*GP*TP*TP*CP*TP*AP*GP*AP*AP*C)-3', ADENOSINE MONOPHOSPHATE, Aprataxin, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2013-10-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity.
Nature, 506, 2013
|
|
4KPE
 
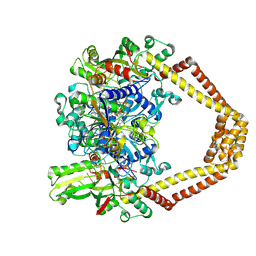 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (7aR,8R)-8-amino-4-cyclopropyl-12-fluoro-1-oxo-4,7,7a,8,9,10-hexahydro-1H-pyrrolo[1',2':1,7]azepino[2,3-h]quinoline-2-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
2IUU
 
 | | P. aeruginosa FtsK motor domain, hexamer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA TRANSLOCASE FTSK | | Authors: | Massey, T.H, Mercogliano, C.P, Yates, J, Sherratt, D.J, Lowe, J. | | Deposit date: | 2006-06-07 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Double-Stranded DNA Translocation: Structure and Mechanism of Hexameric Ftsk
Mol.Cell, 23, 2006
|
|
7NB0
 
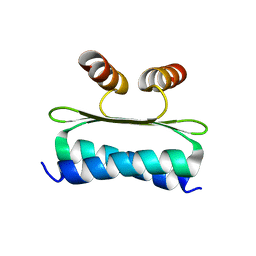 | | Structure of the DNA-binding domain of SEPALLATA 3 | | Descriptor: | Developmental protein SEPALLATA 3 | | Authors: | Zubieta, C, Nanao, M.H. | | Deposit date: | 2021-01-25 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The intervening domain is required for DNA-binding and functional identity of plant MADS transcription factors.
Nat Commun, 12, 2021
|
|
3V7J
 
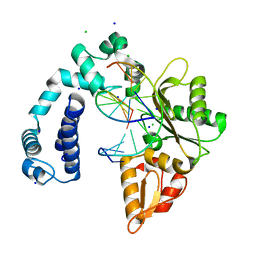 | | Co-crystal structure of Wild Type Rat polymerase beta: Enzyme-DNA binary complex | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*TP*GP*TP*GP*AP*GP*T)-3'), DNA (5'-D(P*CP*AP*AP*AP*CP*TP*CP*AP*CP*AP*TP*A)-3'), ... | | Authors: | Rangarajan, S, Jaeger, J. | | Deposit date: | 2011-12-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic studies of K72E mutant DNA polymerase explain loss of lyase function and reveal changes in the overall conformational state of the polymerase domain
To be Published
|
|
128D
 
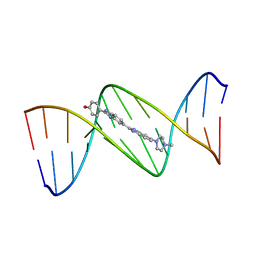 | | MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG) COMPLEXED WITH HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
130D
 
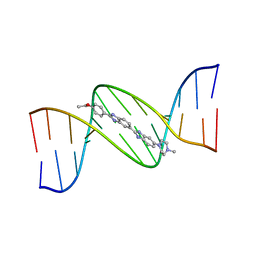 | | MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG) COMPLEXED WITH HOECHST 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
2JXH
 
 | |
7N3Z
 
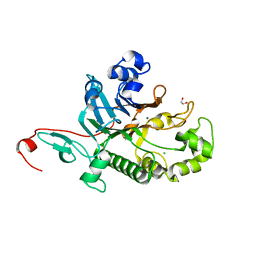 | | Crystal Structure of Saccharomyces cerevisiae Apn2 Catalytic Domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA-(apurinic or apyrimidinic site) endonuclease 2, ... | | Authors: | Wojtaszek, J.L, Wallace, B.D, Williams, R.S. | | Deposit date: | 2021-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular basis for processing of topoisomerase 1-triggered DNA damage by Apn2/APE2.
Cell Rep, 41, 2022
|
|
1JFI
 
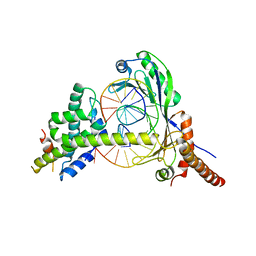 | | Crystal Structure of the NC2-TBP-DNA Ternary Complex | | Descriptor: | 5'-D(*G*GP*AP*GP*CP*CP*CP*TP*TP*TP*TP*AP*TP*AP*GP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*CP*TP*AP*TP*AP*AP*AP*AP*GP*GP*GP*CP*TP*CP*C)-3', TATA-BOX-BINDING PROTEIN (TBP), ... | | Authors: | Kamada, K, Shu, F, Chen, H, Malik, S, Stelzer, G, Roeder, R.G, Meisterernst, M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2001-06-20 | | Release date: | 2001-07-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex.
Cell(Cambridge,Mass.), 106, 2001
|
|
6SEH
 
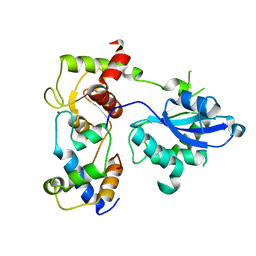 | | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease | | Descriptor: | Structure-specific endonuclease subunit SLX1, Structure-specific endonuclease subunit SLX4, ZINC ION | | Authors: | Gaur, V, Zajko, W, Nirwal, S, Szlachcic, A, Gapinska, M, Nowotny, M. | | Deposit date: | 2019-07-30 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition and processing of branched DNA substrates by Slx1-Slx4 nuclease.
Nucleic Acids Res., 47, 2019
|
|
1JK2
 
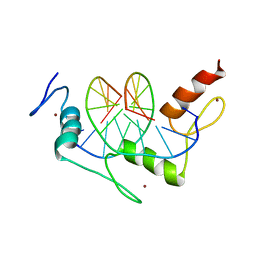 | | Zif268 D20A mutant bound to the GCT DNA site | | Descriptor: | 5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*CP*TP*G)-3', 5'-D(*TP*CP*AP*GP*CP*CP*CP*AP*CP*GP*C)-3', ZIF268, ... | | Authors: | Miller, J.C, Pabo, C.O. | | Deposit date: | 2001-07-11 | | Release date: | 2001-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rearrangement of side-chains in a Zif268 mutant highlights the complexities of zinc finger-DNA recognition.
J.Mol.Biol., 313, 2001
|
|
5AGU
 
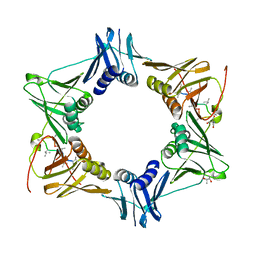 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA POLYMERASE III SUBUNIT BETA, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5AH4
 
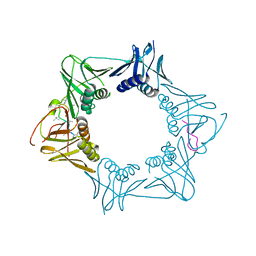 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | CYCLOHEXYL GRISELIMYCIN, DNA POLYMERASE III SUBUNIT BETA, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
8PNC
 
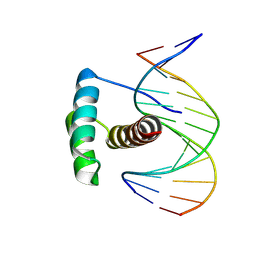 | |
8PMV
 
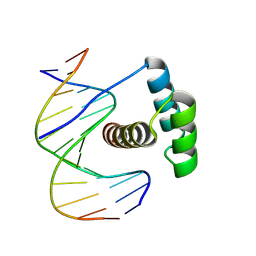 | |
7QW7
 
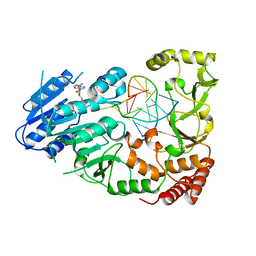 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate fully methylated DNA duplex | | Descriptor: | Fully methylated DNA duplex, Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
7QW5
 
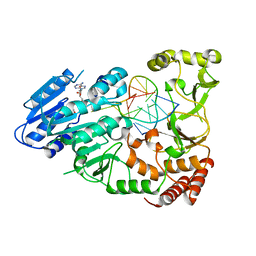 | | Adenine-specific DNA methyltransferase M.BseCI complexed with AdoHcy and cognate unmethylated DNA duplex | | Descriptor: | Modification methylase BseCI, S-ADENOSYL-L-HOMOCYSTEINE, Unmethylated DNA duplex | | Authors: | Mitsikas, D.A, Kouyianou, K, Kotsifaki, D, Providaki, M, Bouriotis, V, Glykos, N.M, Kokkinidis, M. | | Deposit date: | 2022-01-24 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of M.BseCI DNA methyltransferase from Geobacillus stearothermophilus.
To Be Published
|
|
