3MYJ
 
 | |
2AYV
 
 | | Crystal structure of a putative ubiquitin-conjugating enzyme E2 from Toxoplasma gondii | | Descriptor: | UNKNOWN ATOM OR ION, ubiquitin-conjugating enzyme E2 | | Authors: | Tempel, W, Dong, A, Zhao, Y, Lew, J, Alam, Z, Melone, M, Wasney, G, Kozieradzki, I, Vedadi, M, Arrowsmith, C, Sundstrom, M, Weigelt, J, Edwards, A, Bochkarev, A, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-08 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2NYH
 
 | |
3MXF
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective inhibition of BET bromodomains.
Nature, 468, 2010
|
|
3MV8
 
 | | Crystal Structure of the TK3-Gln55His TCR in complex with HLA-B*3501/HPVG | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-35 alpha chain, ... | | Authors: | Gras, S, Chen, Z, Miles, J.J, Liu, Y.C, Bell, M.J, Sullivan, L.C, Kjer-Nielsen, L, Brennan, R.M, Burrows, J.M, Neller, M.A, Khanna, R, Purcell, A.W, Brooks, A.G, McCluskey, J, Rossjohn, J, Burrows, S.R. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-09 | | Last modified: | 2011-07-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Allelic polymorphism in the T cell receptor and its impact on immune responses
J.Exp.Med., 207, 2010
|
|
3MVB
 
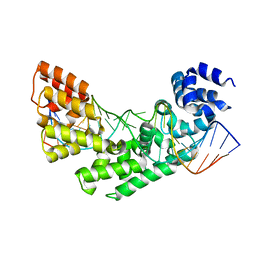 | | Crystal structure of a triple RFY mutant of human MTERF1 bound to the termination sequence | | Descriptor: | 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', 5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', Transcription termination factor, ... | | Authors: | Yakubovskaya, E, Mejia, E, Byrnes, J, Hambardjieva, E, Garcia-Diaz, M. | | Deposit date: | 2010-05-03 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.786 Å) | | Cite: | Helix unwinding and base flipping enable human MTERF1 to terminate mitochondrial transcription.
Cell(Cambridge,Mass.), 141, 2010
|
|
3MVV
 
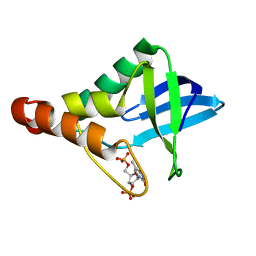 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS F34A at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Caro, J.A, Schlessman, J.L, Garcia-Moreno, E.B, Heroux, A. | | Deposit date: | 2010-05-04 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Cavities determine the pressure unfolding of proteins.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MX6
 
 | |
3MXU
 
 | |
2O1Q
 
 | |
3MV6
 
 | |
2O2G
 
 | |
3MYT
 
 | | Crystal structure of Ketosteroid Isomerase D38HD99N from Pseudomonas testosteroni (tKSI) | | Descriptor: | EQUILENIN, GLYCEROL, SULFATE ION, ... | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2010-05-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Crystal Structure of Ketosteroid Isomerase D38HD99N from Pseudomonas Testosteroni (tKSI)
To be Published
|
|
3MV4
 
 | |
3MWF
 
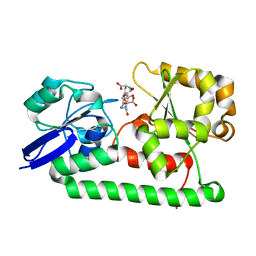 | | Crystal structure of Staphylococcus aureus SirA complexed with staphyloferrin B | | Descriptor: | 5-[(2-{[(3S)-5-{[(2S)-2-amino-2-carboxyethyl]amino}-3-carboxy-3-hydroxy-5-oxopentanoyl]amino}ethyl)amino]-2,5-dioxopentanoic acid, FE (III) ION, Iron-regulated ABC transporter siderophore-binding protein SirA | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Staphylococcus aureus SirA specificity for staphyloferrin B is driven by localized conformational change
To be Published
|
|
3N4W
 
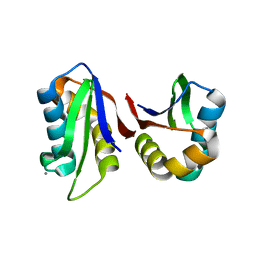 | | Crystal structure of an abridged SER to ALA mutant of the mature ectodomain of the human receptor-type protein-tyrosine phosphatase ICA512/IA-2 at pH 7.5 | | Descriptor: | CALCIUM ION, Receptor-type tyrosine-protein phosphatase-like N | | Authors: | Primo, M.E, Jakoncic, J, Poskus, E, Ermacora, M.R. | | Deposit date: | 2010-05-23 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the mature ectodomain of the human receptor-type protein-tyrosine phosphatase IA-2.
J.Biol.Chem., 283, 2008
|
|
3N56
 
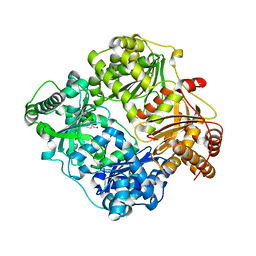 | |
2GHP
 
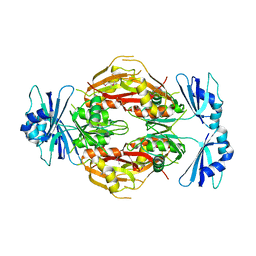 | | Crystal structure of the N-terminal 3 RNA binding domains of the yeast splicing factor Prp24 | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
2NVN
 
 | |
2NUX
 
 | |
3NHD
 
 | | GYVLGS segment 127-132 from human prion with V129 | | Descriptor: | ACETIC ACID, Major prion protein | | Authors: | Apostol, M.I, Eisenberg, D. | | Deposit date: | 2010-06-14 | | Release date: | 2010-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystallographic studies of prion protein (PrP) segments suggest how structural changes encoded by polymorphism at residue 129 modulate susceptibility to human prion disease.
J.Biol.Chem., 285, 2010
|
|
3NI8
 
 | | Crystal Structure of PFC0360w, an HSP90 activator from plasmodium falciparum | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PFC0360w protein | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Pizzaro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PFC0360w, an HSP90 activator from plasmodium falciparum
To be Published
|
|
3NIZ
 
 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rhodanese family protein | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with ADP bound.
To be Published
|
|
3NJG
 
 | | K98A mutant of SO1698 protein, an aspartic peptidase from Shewanella oneidensis. | | Descriptor: | Peptidase | | Authors: | Osipiuk, J, Mulligan, R, Bargassa, M, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Characterization of member of DUF1888 protein family, self-cleaving and self-assembling endopeptidase.
J.Biol.Chem., 287, 2012
|
|
3NJQ
 
 | |
