3ND3
 
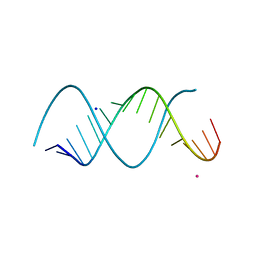 | | Uhelix 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*UP*UP*UP*UP*UP*UP*U)-3', POTASSIUM ION, SODIUM ION | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
3F4H
 
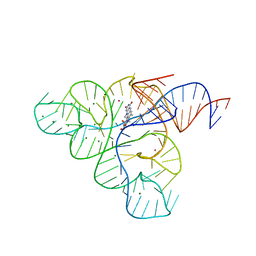 | | Crystal structure of the FMN riboswitch bound to roseoflavin | | Descriptor: | 1-deoxy-1-[8-(dimethylamino)-7-methyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl]-D-ribitol, FMN riboswitch, MAGNESIUM ION, ... | | Authors: | Serganov, A.A, Huang, L. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch.
Nature, 458, 2009
|
|
3F2Q
 
 | |
3F31
 
 | | Crystal Structure of the N-terminal region of AlphaII-spectrin Tetramerization Domain | | Descriptor: | Spectrin alpha chain, brain | | Authors: | Mehboob, S, Santarsiero, B.D, Long, F, Witek, M, Fung, L.W. | | Deposit date: | 2008-10-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the nonerythroid alpha-spectrin tetramerization site reveals differences between erythroid and nonerythroid spectrin tetramer formation.
J.Biol.Chem., 285, 2010
|
|
3F4G
 
 | |
3F2X
 
 | |
3F2Y
 
 | |
3ND4
 
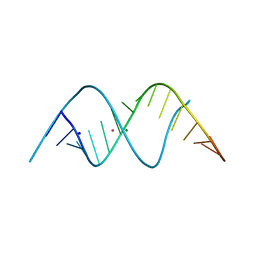 | | Watson-Crick 16-mer dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*GP*AP*AP*GP*AP*UP*CP*UP*UP*CP*UP*CP*U)-3', MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Mooers, B.H, Singh, A. | | Deposit date: | 2010-06-07 | | Release date: | 2011-09-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.524 Å) | | Cite: | The crystal structure of an oligo(U):pre-mRNA duplex from a trypanosome RNA editing substrate.
Rna, 17, 2011
|
|
3FCS
 
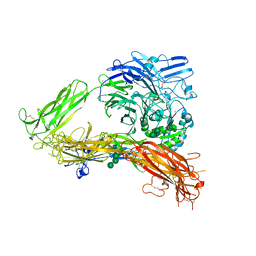 | | Structure of complete ectodomain of integrin aIIBb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Zhu, J, Luo, B.-H, Xiao, T, Zhang, C, Nishida, N, Springer, T.A. | | Deposit date: | 2008-11-22 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces.
Mol.Cell, 32, 2008
|
|
3F30
 
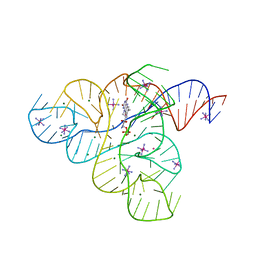 | |
3F2T
 
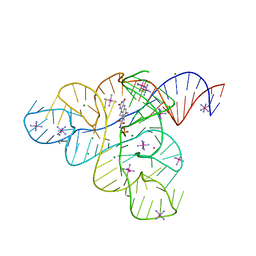 | |
4CAK
 
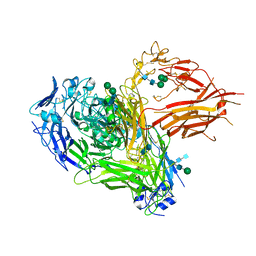 | | Three-dimensional reconstruction of intact human integrin alphaIIbbeta3 in a phospholipid bilayer nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-IIb, ... | | Authors: | Choi, W.S, Rice, W.J, Stokes, D.L, Coller, B.S. | | Deposit date: | 2013-10-08 | | Release date: | 2013-10-30 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (20.5 Å) | | Cite: | Three-Dimensional Reconstruction of Intact Human Integrin Alphaiibbeta3; New Implications for Activation-Dependent Ligand Binding.
Blood, 122, 2013
|
|
7CMD
 
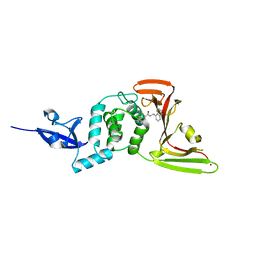 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CJD
 
 | |
3F4E
 
 | |
4DID
 
 | |
8Y1R
 
 | | in situ room temperature Laue crystallography | | Descriptor: | Lysozyme C | | Authors: | Wang, Z.J, Wang, S.S, Pan, Q.Y, Yu, L, Su, Z.H, Yang, T.Y, Wang, Y.Z, Zhang, W.Z, Hao, Q, Gao, X.Y. | | Deposit date: | 2024-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Single pulse data collection with an X-ray chopper at in situ room temperature Laue crystallography beamline BL03HB
Nucl Instrum Methods Phys Res A, 1069, 2024
|
|
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
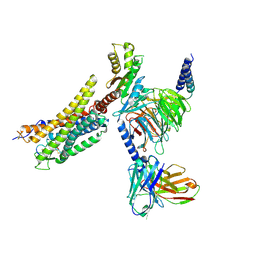 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
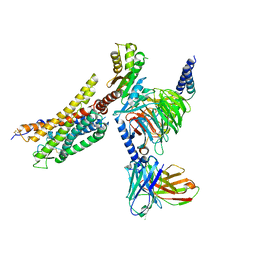 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
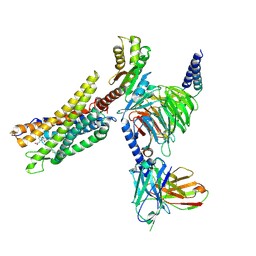 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
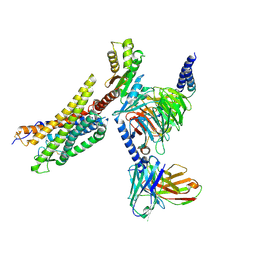 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8ZJV
 
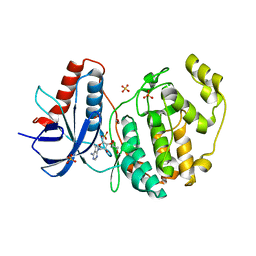 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
7P1P
 
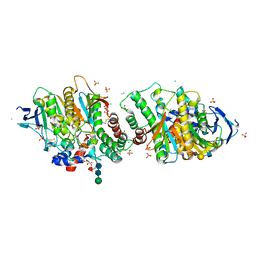 | | Crystal structure of human acetylcholinesterase in complex with (E)-3-hydroxy-6-(3-(4-(4-(((2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)oxy)butyl)-1H-1,2,3-triazol-1-yl)propyl)picolinaldehyde oxime | | Descriptor: | (2R,3R,4S,5S,6R)-2-[4-[1-[3-[6-[(Z)-hydroxyiminomethyl]-5-oxidanyl-pyridin-2-yl]propyl]-1,2,3-triazol-4-yl]butoxy]-6-(hydroxymethyl)oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Ophelie, D.S, Jose, D, Florian, N. | | Deposit date: | 2021-07-02 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A New Class of Bi- and Trifunctional Sugar Oximes as Antidotes against Organophosphorus Poisoning.
J.Med.Chem., 65, 2022
|
|
8YAX
 
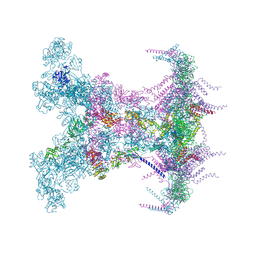 | | SARS-CoV-2 DMV nsp3-4 pore complex (full-pore) | | Descriptor: | Non-structural protein 4, Papain-like protease nsp3 | | Authors: | Huang, Y.X, Zhong, L.J, Zhang, W.X, Ni, T. | | Deposit date: | 2024-02-10 | | Release date: | 2024-06-19 | | Last modified: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Molecular architecture of coronavirus double-membrane vesicle pore complex.
Nature, 633, 2024
|
|
