5ISY
 
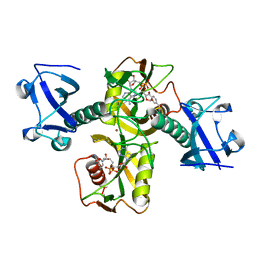 | | Crystal structure of Nudix family protein with NAD | | Descriptor: | NADH pyrophosphatase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-15 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.354 Å) | | Cite: | Structural basis of prokaryotic NAD-RNA decapping by NudC
Cell Res., 26, 2016
|
|
7TZU
 
 | |
7TZS
 
 | |
7TZT
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with N1,N1-dimethyl-N2-(quinoxalin-6-ylmethyl)ethane-1,2-diamine (linked compound 37) | | Descriptor: | 6-methylquinoxaline, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Nuthanakanti, A, Serganov, A. | | Deposit date: | 2022-02-16 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | SHAPE-enabled fragment-based ligand discovery for RNA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7TZR
 
 | |
1A79
 
 | |
5JXS
 
 | |
1OSW
 
 | | The Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop | | Descriptor: | 5'-R(*GP*GP*AP*GP*GP*CP*GP*CP*UP*AP*CP*GP*GP*CP*GP*AP*GP*GP*CP*UP*CP*CP*A)-3' | | Authors: | Yuan, Y, Kerwood, D.J, Paoletti, A.C, Shubsda, M.F, Borer, P.N. | | Deposit date: | 2003-03-20 | | Release date: | 2003-05-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Stem of SL1 RNA in HIV-1: Structure and Nucleocapsid Protein Binding for a 1X3 Internal Loop
Biochemistry, 42, 2003
|
|
2I82
 
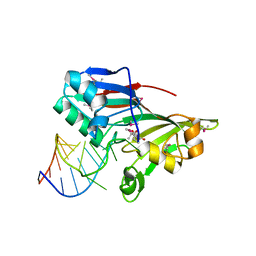 | |
7WV3
 
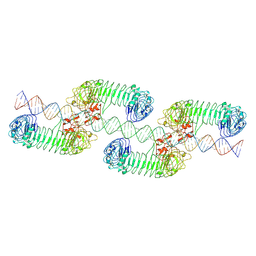 | | Toll-like receptor3 linear cluster | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RNA (80-MER), ... | | Authors: | Lim, C.S, Jang, Y.H, Lee, G.Y, Han, G.M, Lee, J.O. | | Deposit date: | 2022-02-09 | | Release date: | 2022-11-16 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | TLR3 forms a highly organized cluster when bound to a poly(I:C) RNA ligand.
Nat Commun, 13, 2022
|
|
5AXW
 
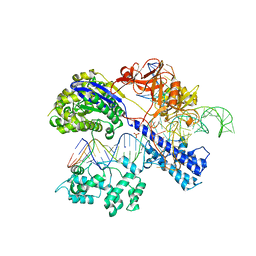 | | Crystal structure of Staphylococcus aureus Cas9 in complex with sgRNA and target DNA (TTGGGT PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, DNA (28-MER), ... | | Authors: | Nishimasu, H, Ishitani, R, Nureki, O. | | Deposit date: | 2015-08-01 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Staphylococcus aureus Cas9.
Cell, 162, 2015
|
|
1L3K
 
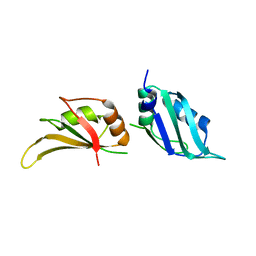 | | UP1, THE TWO RNA-RECOGNITION MOTIF DOMAIN OF HNRNP A1 | | Descriptor: | HETEROGENEOUS NUCLEAR RIBONUCLEOPROTEIN A1 | | Authors: | Vitali, J, Ding, J, Jiang, J, Zhang, Y, Krainer, A.R, Xu, R.-M. | | Deposit date: | 2002-02-27 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Correlated alternative side chain conformations in the RNA-recognition motif of heterogeneous nuclear ribonucleoprotein A1.
Nucleic Acids Res., 30, 2002
|
|
4B3Q
 
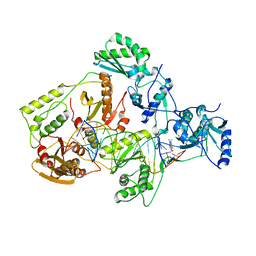 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, P51 RT, PRIMER DNA, ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
8TJV
 
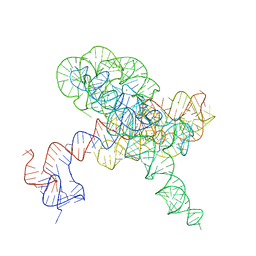 | |
3HKZ
 
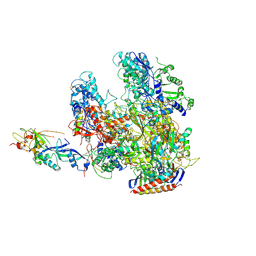 | | The X-ray crystal structure of RNA polymerase from Archaea | | Descriptor: | DNA-directed RNA polymerase subunit 13, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea.
Nature, 451, 2008
|
|
7Z6K
 
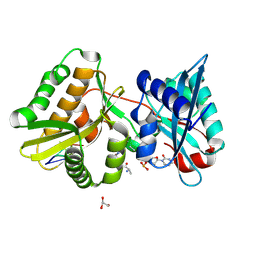 | |
7Z5Z
 
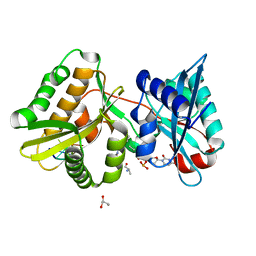 | |
7JJF
 
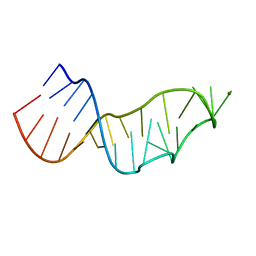 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
4R3Z
 
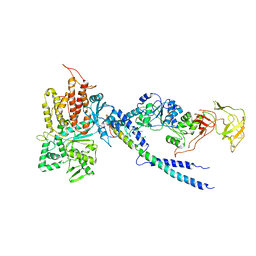 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7L49
 
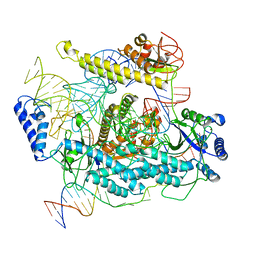 | | Cryo-EM structure of CRISPR-Cas12f Ternary Complex | | Descriptor: | Cas12f1, NTS, Substrate, ... | | Authors: | Chang, L, Li, Z. | | Deposit date: | 2020-12-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for substrate recognition and cleavage by the dimerization-dependent CRISPR-Cas12f nuclease.
Nucleic Acids Res., 49, 2021
|
|
7L3R
 
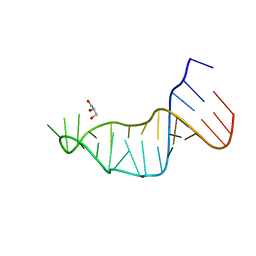 | |
1FKA
 
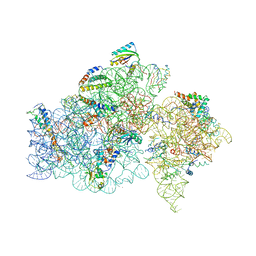 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
8U72
 
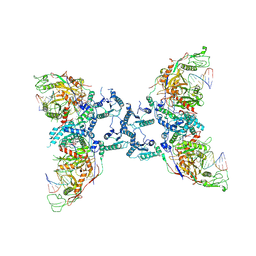 | | Cryo-EM structure of the SPARTA oligomer with guide RNA and target DNA | | Descriptor: | DNA (5'-D(P*TP*AP*TP*AP*CP*AP*AP*CP*CP*TP*AP*CP*TP*AP*CP*CP*TP*C)-3'), MAGNESIUM ION, Piwi domain-containing protein, ... | | Authors: | Malik, R, Kottur, J, Aggarwal, A.K. | | Deposit date: | 2023-09-14 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Nucleic acid mediated activation of a short prokaryotic Argonaute immune system.
Nat Commun, 15, 2024
|
|
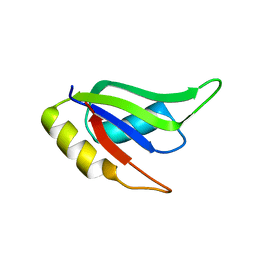
5W0H
 
 | |
5W0G
 
 | | Structure of U2AF65 (U2AF2) RRM1 at 1.07 resolution | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ACETATE ION, Splicing factor U2AF 65 kDa subunit, ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2017-05-30 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Cancer-Associated Mutations Mapped on High-Resolution Structures of the U2AF2 RNA Recognition Motifs.
Biochemistry, 56, 2017
|
|
