3LPO
 
 | | Crystal structure of the N-terminal domain of sucrase-isomaltase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sucrase-isomaltase, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains.
J.Biol.Chem., 285, 2010
|
|
3LPP
 
 | | Crystal complex of N-terminal sucrase-isomaltase with kotalanol | | Descriptor: | (1S,2R,3R,4S)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains.
J.Biol.Chem., 285, 2010
|
|
3LPQ
 
 | | Human MitoNEET with 2Fe-2S Coordinating Ligand His 87 Replaced With Cys | | Descriptor: | CDGSH iron sulfur domain-containing protein 1, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Conlan, A.R, Homer, C, Axelrod, H.L, Cohen, A.E, Abresch, E.C, Zuris, J, Nechushtai, R, Paddock, M.L, Jennings, P.A. | | Deposit date: | 2010-02-05 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mutation of the His ligand in mitoNEET stabilizes the 2Fe-2S cluster despite conformational heterogeneity in the ligand environment.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3LPR
 
 | |
3LPS
 
 | | Crystal structure of parE | | Descriptor: | NOVOBIOCIN, Topoisomerase IV subunit B | | Authors: | Jung, H.Y, Heo, Y.-S. | | Deposit date: | 2010-02-05 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of parE
To be Published
|
|
3LPT
 
 | | HIV integrase | | Descriptor: | (6-chloro-2-oxo-4-phenyl-1,2-dihydroquinolin-3-yl)acetic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
3LPU
 
 | | HIV integrase | | Descriptor: | (2S)-2-(6-chloro-2-methyl-4-phenylquinolin-3-yl)pentanoic acid, 2-[3-[3-(2-hydroxyethoxy)propoxy]propoxy]ethanol, CALCIUM ION, ... | | Authors: | Nicolet, S, Christ, F, Voet, A, Marchand, A, Strelkov, S.V, de Maeyer, M, Chaltin, P, Debyzer, Z. | | Deposit date: | 2010-02-06 | | Release date: | 2010-05-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication.
Nat.Chem.Biol., 6, 2010
|
|
3LPV
 
 | | X-ray crystal structure of duplex DNA containing a cisplatin 1,2-d(GpG) intrastrand cross-link | | Descriptor: | 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', Cisplatin, ... | | Authors: | Todd, R.C, Lippard, S.J. | | Deposit date: | 2010-02-06 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of duplex DNA containing the cisplatin 1,2-{Pt(NH(3))(2)}(2+)-d(GpG) cross-link at 1.77A resolution.
J.Inorg.Biochem., 104, 2010
|
|
3LPW
 
 | | Crystal structure of the FnIII-tandem A77-A78 from the A-band of titin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, A77-A78 domain from Titin | | Authors: | Bucher, R.M, Mayans, O. | | Deposit date: | 2010-02-06 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of the FnIII Tandem A77-A78 points to a periodically conserved architecture in the myosin-binding region of titin
J.Mol.Biol., 401, 2010
|
|
3LPX
 
 | | Crystal structure of GyrA | | Descriptor: | DNA gyrase, A subunit | | Authors: | Jung, H.Y, Heo, Y.S. | | Deposit date: | 2010-02-07 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of GyrA
To be Published
|
|
3LPY
 
 | | Crystal structure of the RRM domain of CyP33 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase E, SULFATE ION | | Authors: | Wang, Z, Patel, D.J. | | Deposit date: | 2010-02-07 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression.
Cell(Cambridge,Mass.), 141, 2010
|
|
3LPZ
 
 | | Crystal structure of C. therm. Get4 | | Descriptor: | Uncharacterized protein | | Authors: | Bozkurt, G, Wild, K, Sinning, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of Get4 reveals an alpha-solenoid fold adapted for multiple interactions in tail-anchored protein biogenesis.
Febs Lett., 584, 2010
|
|
3LQ0
 
 | | Zymogen structure of crayfish astacin metallopeptidase | | Descriptor: | GLYCEROL, ProAstacin, SULFATE ION, ... | | Authors: | Guevara, T, Yiallouros, I, Kappelhoff, R, Bissdorf, S, Stocker, W, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Proenzyme structure and activation of astacin metallopeptidase
J.Biol.Chem., 285, 2010
|
|
3LQ1
 
 | | Crystal structure of 2-succinyl-6-hydroxy-2,4-cyclohexadiene 1-carboxylic acid synthase/2-oxoglutarate decarboxylase FROM Listeria monocytogenes str. 4b F2365 | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Hu, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Sephchc Synthase from Listeria Monocytogenes
To be Published
|
|
3LQ2
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with low TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQ3
 
 | | Crystal structure of human choline kinase beta in complex with phosphorylated hemicholinium-3 and adenosine nucleotide | | Descriptor: | (2S)-2-[4'-({dimethyl[2-(phosphonooxy)ethyl]ammonio}acetyl)biphenyl-4-yl]-2-hydroxy-4,4-dimethylmorpholin-4-ium, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hong, B.S, Tempel, W, Rabeh, W.M, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity.
J.Biol.Chem., 285, 2010
|
|
3LQ4
 
 | | E. coli pyruvate dehydrogenase complex E1 E235A mutant with high TDP concentration | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Furey, W. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Communication between thiamin cofactors in the Escherichia coli pyruvate dehydrogenase complex E1 component active centers: evidence for a "direct pathway" between the 4'-aminopyrimidine N1' atoms.
J.Biol.Chem., 285, 2010
|
|
3LQ5
 
 | | Structure of CDK9/CyclinT in complex with S-CR8 | | Descriptor: | (2S)-2-({9-(1-methylethyl)-6-[(4-pyridin-2-ylbenzyl)amino]-9H-purin-2-yl}amino)butan-1-ol, Cell division protein kinase 9, Cyclin-T1 | | Authors: | Hole, A.J, Endicott, J.A, Baumli, S. | | Deposit date: | 2010-02-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | CDK Inhibitors Roscovitine and CR8 Trigger Mcl-1 Down-Regulation and Apoptotic Cell Death in Neuroblastoma Cells
Genes Cancer, 1, 2010
|
|
3LQ6
 
 | |
3LQ7
 
 | | Crystal structure of glutathione s-transferase from agrobacterium tumefaciens str. c58 | | Descriptor: | Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Gilmore, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-08 | | Release date: | 2010-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Agrobacterium Tumefaciens
To be Published
|
|
3LQ8
 
 | | Structure of the kinase domain of c-Met bound to XL880 (GSK1363089) | | Descriptor: | Hepatocyte growth factor receptor, N-(3-fluoro-4-{[6-methoxy-7-(3-morpholin-4-ylpropoxy)quinolin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxamide | | Authors: | Lougheed, J.C, Stout, T.J. | | Deposit date: | 2010-02-08 | | Release date: | 2010-05-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases.
Cancer Res., 69, 2009
|
|
3LQ9
 
 | | Crystal structure of human REDD1, a hypoxia-induced regulator of mTOR | | Descriptor: | DNA-damage-inducible transcript 4 protein | | Authors: | Vega-Rubin-de-Celis, S, Abdallah, Z, Brugarolas, J, Zhang, X. | | Deposit date: | 2010-02-08 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis and functional implications of the negative mTORC1 regulator REDD1.
Biochemistry, 49, 2010
|
|
3LQA
 
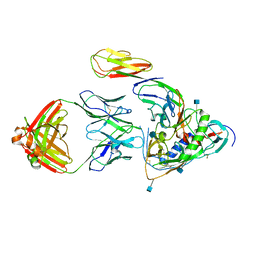 | | Crystal structure of clade C gp120 in complex with sCD4 and 21c Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, Heavy chain of anti HIV Fab from human 21c antibody, ... | | Authors: | Diskin, R, Marcovecchio, P.M, Bjorkman, P.J. | | Deposit date: | 2010-02-08 | | Release date: | 2010-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3LQB
 
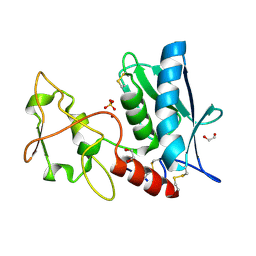 | | Crystal structure of the hatching enzyme ZHE1 from the zebrafish Danio rerio | | Descriptor: | 1,2-ETHANEDIOL, LOC792177 protein, SULFATE ION, ... | | Authors: | Tanokura, M, Okada, A, Nagata, K, Yasumasu, S, Ohtsuka, J, Iuchi, I. | | Deposit date: | 2010-02-08 | | Release date: | 2010-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of zebrafish hatching enzyme 1 from the zebrafish Danio rerio
J.Mol.Biol., 402, 2010
|
|
3LQC
 
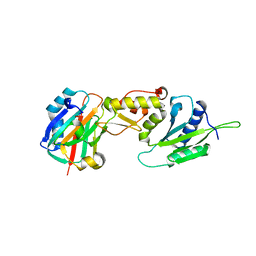 | | X-ray crystal structure of oxidized XRCC1 bound to DNA pol beta Palm thumb domain | | Descriptor: | CARBONATE ION, DNA polymerase beta, DNA repair protein XRCC1, ... | | Authors: | Cuneo, M.J, Krahn, J.M, London, R.E. | | Deposit date: | 2010-02-09 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
