5R94
 
 | |
5R9H
 
 | |
5R9Y
 
 | |
5R95
 
 | | PanDDA analysis group deposition Form1 MAP kinase p38-alpha -- Fragment KCL093 in complex with MAP kinase p38-alpha | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-aminophenyl)pyrrole-2,5-dione, CHLORIDE ION, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
5R9G
 
 | | PanDDA analysis group deposition Form1 MAP kinase p38-alpha -- Fragment PC587 in complex with MAP kinase p38-alpha | | Descriptor: | (4R,4aS,7aS,9S)-3,10-dimethyl-5,6,7,7a,8,9-hexahydro-4H-4a,9-epiminopyrrolo[3',4':5,6]cyclohepta[1,2-d][1,2]oxazol-4-ol, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | De Nicola, G.F, Nichols, C.E. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Mining the PDB for Tractable Cases Where X-ray Crystallography Combined with Fragment Screens Can Be Used to Systematically Design Protein-Protein Inhibitors: Two Test Cases Illustrated by IL1 beta-IL1R and p38 alpha-TAB1 Complexes.
J.Med.Chem., 63, 2020
|
|
5R9X
 
 | |
4W4V
 
 | | JNK2/3 in complex with 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(2-chlorophenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4Y
 
 | | JNK2/3 in complex with 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide | | Descriptor: | 3-(4-{[(4-methylphenyl)carbamoyl]amino}-1H-pyrazol-1-yl)-N-(2-methylpyridin-4-yl)benzamide, c-jun NH2-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
4W4W
 
 | | JNK2/3 in complex with N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide | | Descriptor: | N-(2-methylpyridin-4-yl)-3-{4-[(phenylcarbamoyl)amino]-1H-pyrazol-1-yl}benzamide, c-Jun N-terminal kinase 3 | | Authors: | Park, H, Iqbal, S, Hernandez, P, Mora, R, Zheng, K, Feng, Y, LoGrasso, P. | | Deposit date: | 2014-08-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis and Biological Consequences for JNK2/3 Isoform Selective Aminopyrazoles.
Sci Rep, 5, 2015
|
|
1R3C
 
 | | THE STRUCTURE OF P38ALPHA C162S MUTANT | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
8EFJ
 
 | |
1R39
 
 | | THE STRUCTURE OF P38ALPHA | | Descriptor: | Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Patel, S.B, Cameron, P.M, Frantz-Wattley, B, O'Neill, E, Becker, J.W, Scapin, G. | | Deposit date: | 2003-10-01 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Lattice stabilization and enhanced diffraction in human p38 alpha crystals by protein engineering.
Biochim.Biophys.Acta, 1696, 2004
|
|
3GP0
 
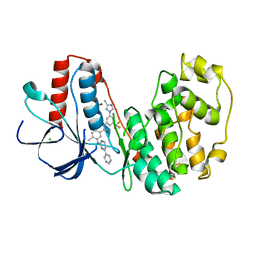 | | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Mitogen-activated protein kinase 11, ... | | Authors: | Filippakopoulos, P, Barr, A, Fedorov, O, Keates, T, Soundararajan, M, Elkins, J, Salah, E, Burgess-Brown, N, Ugochukwu, E, Pike, A.C.W, Muniz, J, Roos, A, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Mitogen Activated Protein Kinase 11 (p38 beta) in complex with Nilotinib
To be Published
|
|
2GFS
 
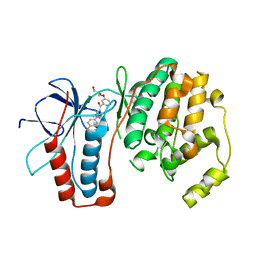 | | P38 Kinase Crystal Structure in complex with RO3201195 | | Descriptor: | Mitogen-Activated Protein Kinase 14, [5-AMINO-1-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL](3-{[(2R)-2,3-DIHYDROXYPROPYL]OXY}PHENYL)METHANONE | | Authors: | Harris, S.F, Bertrand, J, Villasenor, A. | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Discovery of S-[5-Amino-1-(4-fluorophenyl)-1H-pyrazol-4-yl]-[3-(2,3-dihydroxypropoxy)phenyl]-methanone (RO3201195), and Orally Bioavailable and Highly Selective Inhibitor of p38 Map Kinase
J.Med.Chem., 49, 2006
|
|
2GHL
 
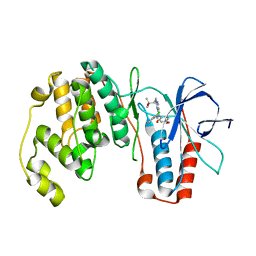 | | Mutant Mus Musculus P38 Kinase Domain in Complex with Inhibitor PG-874743 | | Descriptor: | 3-(2-CHLOROPHENYL)-1-(2-{[(1S)-2-HYDROXY-1,2-DIMETHYLPROPYL]AMINO}PYRIMIDIN-4-YL)-1-(4-METHOXYPHENYL)UREA, Mitogen-activated protein kinase 14 | | Authors: | Walter, R.L, Mekel, M.J, Evdokimov, A.G, Pokross, M.E, Brugel, T.A. | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Development of N-2,4-pyrimidine-N-phenyl-N'-phenyl ureas as inhibitors of tumor necrosis factor alpha (TNF-alpha) synthesis. Part 1.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6DS2
 
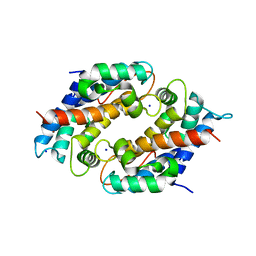 | | Crystal structure of Ni(II)-bound human calprotectin | | Descriptor: | NICKEL (II) ION, Protein S100-A8, Protein S100-A9, ... | | Authors: | Nolan, E.M, Drennan, C.L, Nakashige, T.G. | | Deposit date: | 2018-06-13 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biophysical Examination of the Calcium-Modulated Nickel-Binding Properties of Human Calprotectin Reveals Conformational Change in the EF-Hand Domains and His3Asp Site.
Biochemistry, 57, 2018
|
|
1LEW
 
 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS NUCLEAR SUBSTRATE MEF2A | | Descriptor: | Mitogen-activated protein kinase 14, Myocyte-specific enhancer factor 2A | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
1IRJ
 
 | | Crystal Structure of the MRP14 complexed with CHAPS | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CALCIUM ION, Migration Inhibitory Factor-Related Protein 14 | | Authors: | Itou, H, Yao, M, Watanabe, N, Nishihira, J, Tanaka, I. | | Deposit date: | 2001-10-09 | | Release date: | 2002-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human MRP14 (S100A9), a Ca(2+)-dependent regulator protein in inflammatory process.
J.Mol.Biol., 316, 2002
|
|
3HA8
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/Compound 14b | | Descriptor: | Mitogen-activated protein kinase 14, N~2~-{4-[6-(3,4-dihydroquinolin-1(2H)-ylcarbonyl)-1H-benzimidazol-1-yl]-6-ethoxy-1,3,5-triazin-2-yl}-3-(2,2-dimethyl-4H-1,3-benzodioxin-6-yl)-N-methyl-L-alaninamide | | Authors: | Zhao, B, Clark, M.A. | | Deposit date: | 2009-05-01 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design, synthesis and selection of DNA-encoded small-molecule libraries.
Nat.Chem.Biol., 5, 2009
|
|
3HEC
 
 | | P38 in complex with Imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
3HEG
 
 | | P38 in complex with Sorafenib | | Descriptor: | 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Mitogen-activated protein kinase 14 | | Authors: | Namboodiri, H.V, Karpusas, M. | | Deposit date: | 2009-05-08 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analysis of imatinib and sorafenib binding to p38alpha compared with c-Abl and b-Raf provides structural insights for understanding the selectivity of inhibitors targeting the DFG-out form of protein kinases.
Biochemistry, 49, 2010
|
|
1CWT
 
 | | HUMAN CDC25B CATALYTIC DOMAIN WITH METHYL MERCURY | | Descriptor: | CDC25 B-TYPE TYROSINE PHOSPHATASE, CHLORIDE ION, METHYL MERCURY ION, ... | | Authors: | Watenpaugh, K.D, Reynolds, R.A. | | Deposit date: | 1999-08-26 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle.
J.Mol.Biol., 293, 1999
|
|
1CWS
 
 | | HUMAN CDC25B CATALYTIC DOMAIN WITH TUNGSTATE | | Descriptor: | BETA-MERCAPTOETHANOL, CDC25 B-TYPE TYROSINE PHOSPHATASE, CHLORIDE ION, ... | | Authors: | Watenpaugh, K.D, Reynolds, R.A, Chidester, C.G. | | Deposit date: | 1999-08-26 | | Release date: | 2000-08-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle.
J.Mol.Biol., 293, 1999
|
|
1CWR
 
 | |
7CGA
 
 | |
