7Z9L
 
 | | Phen-DC3 intercalation causes hybrid-to-antiparallel transformation of human telomeric DNA G-quadruplex | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3'), N2,N9-bis(1-methylquinolin-3-yl)-1,10-phenanthroline-2,9-dicarboxamide | | Authors: | Ghosh, A, Trajkovski, M, Teulade-Fichou, M.P, Gabelica, V, Plavec, J. | | Deposit date: | 2022-03-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phen-DC 3 Induces Refolding of Human Telomeric DNA into a Chair-Type Antiparallel G-Quadruplex through Ligand Intercalation.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2LVJ
 
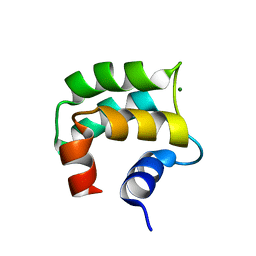 | | solution structure of hemi-Mg-bound Phl p 7 | | Descriptor: | MAGNESIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
3L1Y
 
 | |
5E22
 
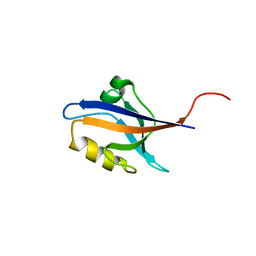 | | The second PDZ domain of Ligand of Numb protein X 2 in the presence of an electric field of ~1 MV/cm along the crystallographic x axis, with eightfold extrapolation of structure factor differences. | | Descriptor: | GLYCEROL, Ligand of Numb protein X 2 | | Authors: | Hekstra, D.R, White, K.I, Socolich, M.A, Henning, R.W, Srajer, V, Ranganathan, R. | | Deposit date: | 2015-09-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Electric-field-stimulated protein mechanics.
Nature, 540, 2016
|
|
7CT6
 
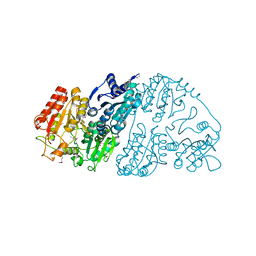 | | Crystal structure of GCL from Deinococcus metallilatus | | Descriptor: | Glyoxylate carboligase | | Authors: | Kim, J.H, Kim, J.S. | | Deposit date: | 2020-08-18 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Glyoxylate carboligase-based whole-cell biotransformation of formaldehyde into ethylene glycol via glycolaldehyde.
Green Chem, 1, 2022
|
|
4XV5
 
 | | CcP gateless cavity | | Descriptor: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
2Y7Y
 
 | | APLYSIA CALIFORNICA ACHBP IN APO STATE | | Descriptor: | SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, De Esch, I.J. | | Deposit date: | 2011-02-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X-Ray Analysis.
J.Med.Chem., 52, 2009
|
|
4XV7
 
 | | CcP gateless cavity | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
3HJO
 
 | |
3HJM
 
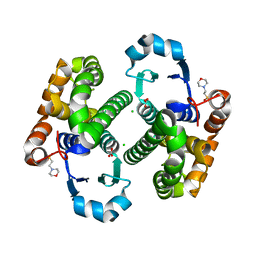 | | Crystal structure of human Glutathione Transferase Pi Y108V mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Parker, L.J. | | Deposit date: | 2009-05-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Influence of the H-site residue 108 on human glutathione transferase P1-1 ligand binding: structure-thermodynamic relationships and thermal stability.
Protein Sci., 18, 2009
|
|
2MOA
 
 | |
7B7N
 
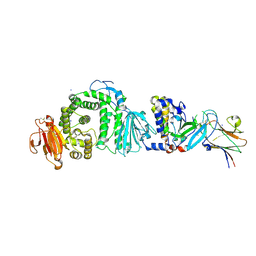 | | Human herpesvirus-8 gH/gL in complex with EphA2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Pederzoli, R, Guardado-Calvo, P, Rey, F.A, Backovic, M. | | Deposit date: | 2020-12-11 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Human herpesvirus 8 molecular mimicry of ephrin ligands facilitates cell entry and triggers EphA2 signaling.
Plos Biol., 19, 2021
|
|
4MG6
 
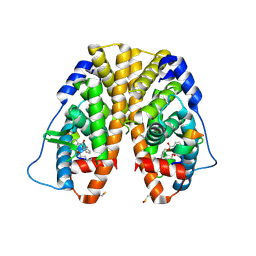 | | Crystal structure of hERa-LBD (Y537S) in complex with benzylbutylphtalate | | Descriptor: | Estrogen receptor, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4D8D
 
 | |
4XV8
 
 | | CcP gateless cavity | | Descriptor: | BENZAMIDINE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
6EU5
 
 | | Leishmania major N-myristoyltransferase with bound myristoyl-CoA and inhibitor | | Descriptor: | 4-[3-[(8~{a}~{R})-3,4,6,7,8,8~{a}-hexahydro-1~{H}-pyrrolo[1,2-a]pyrazin-2-yl]propyl]-2,6-bis(chloranyl)-~{N}-methyl-~{N}-(1,3,5-trimethylpyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA | | Authors: | Brenk, R, Kehrein, J, Kersten, C. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496083 Å) | | Cite: | How To Design Selective Ligands for Highly Conserved Binding Sites: A Case Study UsingN-Myristoyltransferases as a Model System.
J.Med.Chem., 2019
|
|
2LVI
 
 | | Solution structure of apo-Phl p 7 | | Descriptor: | Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
6EWF
 
 | |
4XV6
 
 | | CcP gateless cavity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
3R1H
 
 | | Crystal structure of the Class I ligase ribozyme-substrate preligation complex, C47U mutant, Ca2+ bound | | Descriptor: | 5'-R(*UP*CP*CP*AP*GP*UP*A)-3', CALCIUM ION, Class I ligase ribozyme, ... | | Authors: | Shechner, D.M, Bartel, D.P. | | Deposit date: | 2011-03-10 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The structural basis of RNA-catalyzed RNA polymerization.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2LVK
 
 | | Solution structure of Ca-bound Phl p 7 | | Descriptor: | CALCIUM ION, Polcalcin Phl p 7 | | Authors: | Henzl, M.T, Sirianni, A.G, Tanner, J.J. | | Deposit date: | 2012-07-05 | | Release date: | 2012-10-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of polcalcin Phl p 7 in three ligation states: Apo-, hemi-Mg(2+) -bound, and fully Ca(2+) -bound.
Proteins, 81, 2013
|
|
4XVA
 
 | | Crystal structure of wild type cytochrome c peroxidase | | Descriptor: | BENZIMIDAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
4MGC
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with benzophenone-2 | | Descriptor: | Estrogen receptor, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4MGA
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with 4-tert-octylphenol | | Descriptor: | 4-(2,4,4-trimethylpentan-2-yl)phenol, Estrogen receptor, Nuclear receptor coactivator 1 | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4XV4
 
 | | CcP gateless cavity | | Descriptor: | 2-AMINO-5-METHYLTHIAZOLE, Cytochrome c peroxidase, mitochondrial, ... | | Authors: | Fischer, M, Fraser, J.S. | | Deposit date: | 2015-01-26 | | Release date: | 2015-02-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | One Crystal, Two Temperatures: Cryocooling Penalties Alter Ligand Binding to Transient Protein Sites.
Chembiochem, 16, 2015
|
|
