2BXW
 
 | | CRYSTAL STRUCTURE OF RHOGDI Lys(135,138,141)Tyr MUTANT | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, RHO GDP-DISSOCIATION INHIBITOR 1 | | Authors: | Sikorska, M, Cooper, D.R, Otlewski, J, Derewenda, Z.S. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protein Crystallization by Surface Entropy Reduction: Optimization of the Ser Strategy
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1J17
 
 | |
5SOU
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000285507655 - (R) and (S) isomers | | Descriptor: | 5-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one, 5-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4IBY
 
 | | Human p53 core domain with hot spot mutation R273H and second-site suppressor mutation S240R | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | Authors: | Eldar, A, Rozenberg, H, Diskin-Posner, Y, Shakked, Z. | | Deposit date: | 2012-12-09 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
5RWI
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434926 | | Descriptor: | 4-[(furan-2-yl)methyl]-1lambda~6~,4-thiazinane-1,1-dione, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
4IDG
 
 | | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Vetting, M.W, Groninger-Poe, F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-12 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a short-chain dehydrogenase/reductase superfamily protein from agrobacterium tumefaciens (TARGET EFI-506441) with bound NAD, monoclinic form 2
To be Published
|
|
5SRS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z2614735107 - (R) and (S) isomers | | Descriptor: | 3-[(3R)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, 3-[(3S)-1-(7-fluoro-9H-pyrimido[4,5-b]indol-4-yl)pyrrolidin-3-yl]-1,3-oxazolidin-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2QQ0
 
 | | Thymidine Kinase from Thermotoga Maritima in complex with thymidine + AppNHp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Segura-Pena, D, Lichter, J, Trani, M, Konrad, M, Lavie, A, Lutz, S. | | Deposit date: | 2007-07-25 | | Release date: | 2007-10-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Quaternary structure change as a mechanism for the regulation of thymidine kinase 1-like enzymes.
Structure, 15, 2007
|
|
5PO1
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10152a | | Descriptor: | 1,2-ETHANEDIOL, 5-methyl-N-[(thiophen-2-yl)methyl]-1,2-oxazol-3-amine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5POU
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N10954a | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 1, N-(1-benzylpiperidin-4-yl)-N'-methylurea, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3ZV2
 
 | |
3AL7
 
 | | Recombinant thaumatin I at 1.1 A | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Masuda, T, Mikami, B, Kitabatake, N. | | Deposit date: | 2010-07-27 | | Release date: | 2011-06-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-resolution structure of the recombinant sweet-tasting protein thaumatin I
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6LKR
 
 | | Crystal structure of mouse DCAR2 CRD domain complex | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, C-type lectin domain family 4, member b1, ... | | Authors: | Omahdi, Z, Horikawa, Y, Toyonaga, K, Kakuta, Y, Yamasaki, S. | | Deposit date: | 2019-12-20 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insight into the recognition of pathogen-derived phosphoglycolipids by C-type lectin receptor DCAR.
J.Biol.Chem., 295, 2020
|
|
6B95
 
 | | Multiconformer model of K197C PTP1B tethered to compound 2 at 100 K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(2',4'-difluoro-4-hydroxy[1,1'-biphenyl]-3-yl)-2-sulfanylacetamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4KQF
 
 | | Crystal structure of CobT E174A complexed with adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, ... | | Authors: | Chan, C.H, Newmister, S.A, Taylor, K.C, Claas, K.R, Rayment, I, Escalante-Semerena, J.C. | | Deposit date: | 2013-05-15 | | Release date: | 2014-03-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dissecting cobamide diversity through structural and functional analyses of the base-activating CobT enzyme of Salmonella enterica.
Biochim.Biophys.Acta, 1840, 2014
|
|
5SPC
 
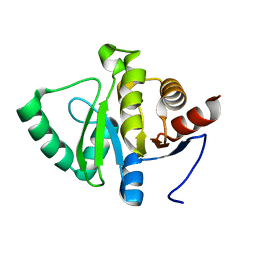 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894387 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3PA4
 
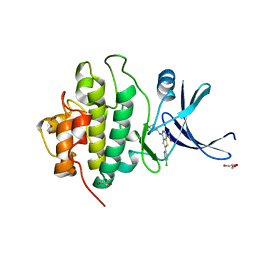 | | X-ray crystal structure of compound 2a bound to human CHK1 kinase domain | | Descriptor: | 2-(4-chlorophenyl)-4-[(3S)-piperidin-3-ylamino]thieno[3,2-c]pyridine-7-carboxamide, GLYCEROL, Serine/threonine-protein kinase Chk1 | | Authors: | Fischmann, T.O. | | Deposit date: | 2010-10-18 | | Release date: | 2010-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Design, synthesis and SAR of thienopyridines as potent CHK1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3ZNU
 
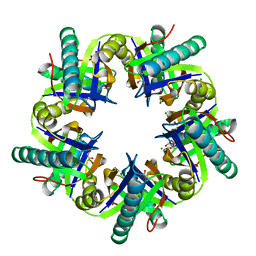 | | Crystal structure of ClcF in crystal form 2 | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION, ... | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-18 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3ZOV
 
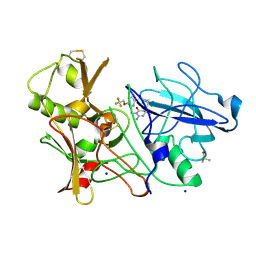 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-02-25 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3GB8
 
 | | Crystal structure of CRM1/Snurportin-1 complex | | Descriptor: | Exportin-1, Snurportin-1 | | Authors: | Dong, X, Biswas, A, Suel, K.E, Jackson, L.K, Martinez, R, Gu, H, Chook, Y.M. | | Deposit date: | 2009-02-19 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for leucine-rich nuclear export signal recognition by CRM1.
Nature, 458, 2009
|
|
3R0D
 
 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Kamat, S, Hitchcock, D, Raushel, F.M, Almo, S.C. | | Deposit date: | 2011-03-07 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with two zinc atoms in the active site
To be Published
|
|
3RRP
 
 | |
3FUD
 
 | |
5Q1V
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000166a | | Descriptor: | 1-(4-fluorophenyl)-~{N}-[[(2~{R})-oxolan-2-yl]methyl]methanamine, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
2BKF
 
 | |
