8JXL
 
 | |
7YH6
 
 | | Structure of SARS-CoV-2 spike RBD in complex with neutralizing antibody NIV-8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, NIV-8 Fab light chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-12 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
6R6D
 
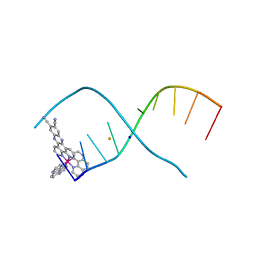 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(TCGGCGCCGA)2 | | Descriptor: | BARIUM ION, DNA (5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3'), Ruthenium (bis-(tetraazaphenanthrene)) (11,12-dicyano-dipyridophenazine), ... | | Authors: | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | Deposit date: | 2019-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
8K2V
 
 | |
5FPA
 
 | |
7M32
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Uracil and NADPH Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ALANINE, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
7MTX
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with IMP and the inhibitor P176 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-{2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenyl}-beta-D-ribopyranosylamine, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-05-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with IMP and the inhibitor P176
To Be Published
|
|
2X86
 
 | | AGME bound to ADP-B-mannose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADP-L-GLYCERO-D-MANNO-HEPTOSE-6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kowatz, T, Morrison, J.P, Tanner, M.E, Naismith, J.H. | | Deposit date: | 2010-03-06 | | Release date: | 2010-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Crystal Structure of the Y140F Mutant of Adp-L-Glycero-D-Manno-Heptose 6-Epimerase Bound to Adp-Beta-D-Mannose Suggests a One Base Mechanism.
Protein Sci., 19, 2010
|
|
6F3G
 
 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | Interleukin-1 receptor-associated kinase 4, SULFATE ION, ~{N}4,~{N}4-dimethyl-~{N}1-(5-propan-2-ylpyrrolo[3,2-d]pyrimidin-4-yl)cyclohexane-1,4-diamine | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
6UIZ
 
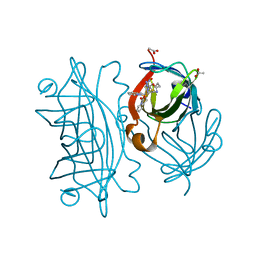 | | Artificial Iron Proteins: Modelling the Active Sites in Non-Heme Dioxygenases | | Descriptor: | ACETATE ION, Streptavidin, {N-(2-{bis[(pyridin-2-yl-kappaN)methyl]amino-kappaN}ethyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(triaza-1,2-dien-2-ium-1-ide-kappaN~1~)iron(4+) | | Authors: | Miller, K.R, Paretsky, J.D, Follmer, A.H, Heinisch, T, Mittra, K, Gul, S, Kim, I.-S, Fuller, F.D, Batyuk, A, Sutherlin, K.D, Brewster, A.S, Bhowmick, A, Sauter, N.K, Kern, J, Yano, J, Green, M.T, Ward, T.R, Borovik, A.S. | | Deposit date: | 2019-10-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Artificial Iron Proteins: Modeling the Active Sites in Non-Heme Dioxygenases.
Inorg.Chem., 59, 2020
|
|
6HKQ
 
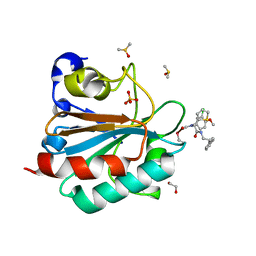 | | Human GPX4 in complex with covalent Inhibitor ML162 (S enantiomer) | | Descriptor: | (2~{S})-2-[2-chloranylethanoyl-(3-chloranyl-4-methoxy-phenyl)amino]-~{N}-(2-phenylethyl)-2-thiophen-2-yl-ethanamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Hillig, R.C, Moosmayer, D, Hilpmann, A, Hoffmann, L, Schnirch, L, Eaton, J.K, Badock, V, Gradl, S. | | Deposit date: | 2018-09-07 | | Release date: | 2020-04-01 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of the selenoprotein glutathione peroxidase 4 in its apo form and in complex with the covalently bound inhibitor ML162.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7XY4
 
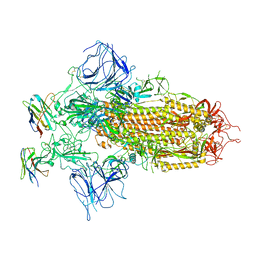 | |
7XY3
 
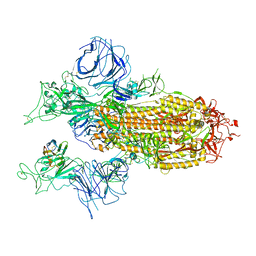 | |
6F3D
 
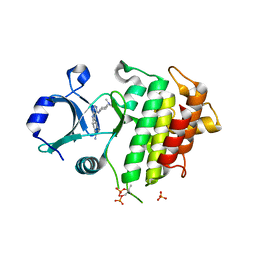 | | IRAK4 IN COMPLEX WITH inhibitor | | Descriptor: | 4-[4-[[4-(dimethylamino)cyclohexyl]amino]-7~{H}-pyrrolo[2,3-d]pyrimidin-5-yl]cyclohexane-1-carboxamide, Interleukin-1 receptor-associated kinase 4, SULFATE ION | | Authors: | Xue, Y, Degorce, S.L, Robb, G.R, Ferguson, A.D. | | Deposit date: | 2017-11-28 | | Release date: | 2018-05-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Optimization of permeability in a series of pyrrolotriazine inhibitors of IRAK4.
Bioorg. Med. Chem., 26, 2018
|
|
5UW4
 
 | |
2XFE
 
 | | vCBM60 in complex with galactobiose | | Descriptor: | CALCIUM ION, CARBOHYDRATE BINDING MODULE, beta-D-galactopyranose-(1-4)-beta-D-galactopyranose | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D.P, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules.
J.Biol.Chem., 285, 2010
|
|
4WMF
 
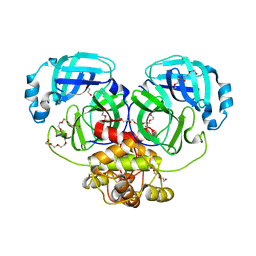 | | Crystal structure of catalytically inactive MERS-CoV 3CL protease (C148A) in spacegroup P212121 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MERS-CoV 3CL protease, TETRAETHYLENE GLYCOL | | Authors: | Lountos, G.T, Needle, D, Waugh, D.S. | | Deposit date: | 2014-10-08 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of the Middle East respiratory syndrome coronavirus 3C-like protease reveal insights into substrate specificity.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XJR
 
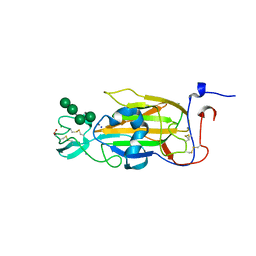 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D2-D3) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5KKN
 
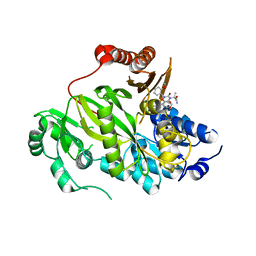 | | Crystal structure of human ACC2 BC domain in complex with ND-646, the primary amide of ND-630 | | Descriptor: | 2-[1-[(2~{R})-2-(2-methoxyphenyl)-2-(oxan-4-yloxy)ethyl]-5-methyl-6-(1,3-oxazol-2-yl)-2,4-bis(oxidanylidene)thieno[2,3-d]pyrimidin-3-yl]-2-methyl-propanamide, Acetyl-CoA carboxylase 2 | | Authors: | Wang, R, Paul, D, Tong, L. | | Deposit date: | 2016-06-22 | | Release date: | 2016-07-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Acetyl-CoA carboxylase inhibition by ND-630 reduces hepatic steatosis, improves insulin sensitivity, and modulates dyslipidemia in rats.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2XJT
 
 | | X-ray structure of the N-terminal domain of the flocculin Flo5 from Saccharomyces cerevisiae in complex with calcium and Man5(D1) | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLOCCULATION PROTEIN FLO5, ... | | Authors: | Veelders, M, Brueckner, S, Ott, D, Unverzagt, C, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2010-07-06 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Flocculin-Mediated Social Behavior in Yeast
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6B7H
 
 | | Structure of mGluR3 with an agonist | | Descriptor: | (1S,2S,4S,5R,6S)-2-amino-4-[(3-methoxybenzene-1-carbonyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Monn, J.A, Clawson, D.K. | | Deposit date: | 2017-10-04 | | Release date: | 2018-04-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Synthesis and Pharmacological Characterization of C4beta-Amide-Substituted 2-Aminobicyclo[3.1.0]hexane-2,6-dicarboxylates. Identification of (1 S,2 S,4 S,5 R,6 S)-2-Amino-4-[(3-methoxybenzoyl)amino]bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid (LY2794193), a Highly Potent and Selective mGlu3Receptor Agonist.
J. Med. Chem., 61, 2018
|
|
2XLL
 
 | | The crystal structure of bilirubin oxidase from Myrothecium verrucaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIRUBIN OXIDASE, COPPER (II) ION | | Authors: | McNamara, T.P, Lowe, E.D, Cracknell, J.A, Blanford, C.F. | | Deposit date: | 2010-07-21 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Bilirubin Oxidase from Myrothecium Verrucaria: X- Ray Determination of the Complete Crystal Structure and a Rational Surface Modification for Enhanced Electrocatalytic O(2) Reduction.
Dalton Trans, 40, 2011
|
|
2XQK
 
 | | X-ray Structure of human butyrylcholinesterase inhibited by pure enantiomer VX-(S) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wandhammer, M, Carletti, E, Gillon, E, Masson, P, Goeldner, M, Noort, D, Nachon, F. | | Deposit date: | 2010-09-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Study of the Complex Stereoselectivity of Human Butyrylcholinesterase for the Neurotoxic V-Agents.
J.Biol.Chem., 286, 2011
|
|
5UN1
 
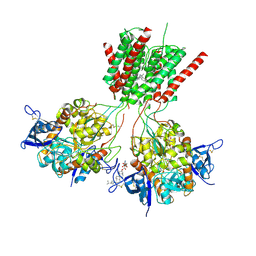 | | Crystal structure of GluN1/GluN2B delta-ATD NMDA receptor | | Descriptor: | (5S,10R)-5-methyl-10,11-dihydro-5H-5,10-epiminodibenzo[a,d][7]annulene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Song, X, Gouaux, E. | | Deposit date: | 2017-01-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of NMDA receptor channel block by MK-801 and memantine.
Nature, 556, 2018
|
|
2XQF
 
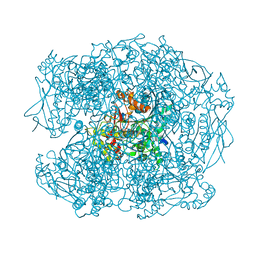 | | X-ray Structure of human butyrylcholinesterase inhibited by racemic VX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Wandhammer, M, Carletti, E, Gillon, E, Masson, P, Goeldner, M, Noort, D, Nachon, F. | | Deposit date: | 2010-09-02 | | Release date: | 2011-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Study of the Complex Stereoselectivity of Human Butyrylcholinesterase for the Neurotoxic V-Agents.
J.Biol.Chem., 286, 2011
|
|
