3B1Q
 
 | | Structure of Burkholderia thailandensis nucleoside kinase (BthNK) in complex with inosine | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ACETATE ION, INOSINE, ... | | Authors: | Yasutake, Y, Ota, H, Hino, E, Sakasegawa, S, Tamura, T. | | Deposit date: | 2011-07-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Burkholderia thailandensis nucleoside kinase: implications for the catalytic mechanism and nucleoside selectivity
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4LBB
 
 | |
3MOZ
 
 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,2,3,5,6)pentakisphosphate | | Descriptor: | (1R,2R,3R,4R,5S,6S)-6-HYDROXYCYCLOHEXANE-1,2,3,4,5-PENTAYL PENTAKIS[DIHYDROGEN (PHOSPHATE)], ACETATE ION, CHLORIDE ION, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-04-23 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate binding in protein-tyrosine phosphatase-like inositol polyphosphatases.
J.Biol.Chem., 287, 2012
|
|
3PBY
 
 | | Crystal structure of the mutant L123S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
1NFX
 
 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR208944 | | Descriptor: | 4-[(6-CHLORO-1-BENZOTHIEN-2-YL)SULFONYL]-1-{[1-(2-HYDROXYETHYL)-1H-PYRROLO[3,2-C]PYRIDIN-2-YL]METHYL}PIPERAZIN-2-ONE, CALCIUM ION, COAGULATION FACTOR XA, ... | | Authors: | Maignan, S, Guilloteau, J.P. | | Deposit date: | 2002-12-16 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular structures of human Factor Xa complexed with ketopiperazine inhibitors:
preference for a neutral group in the S1 pocket.
J.Med.Chem., 46, 2003
|
|
7HJ4
 
 | |
3BAF
 
 | | Crystal structure of shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Shikimate kinase | | Authors: | Faim, L.M, Dias, M.V.B, Vasconcelos, I.G, Basso, L.A, Santos, D.S, Azevedo, W.F, Ruggiero, N.J. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure for shikimate kinase from Mycobacterium tuberculosis in complex with AMP-PNP
To be Published
|
|
3PDY
 
 | |
1VSE
 
 | | ASV INTEGRASE CORE DOMAIN WITH MG(II) COFACTOR AND HEPES LIGAND, LOW MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
1VSF
 
 | | ASV INTEGRASE CORE DOMAIN WITH MN(II) COFACTOR AND HEPES LIGAND, HIGH MG CONCENTRATION FORM | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, INTEGRASE, MANGANESE (II) ION | | Authors: | Bujacz, G, Jaskolski, M, Alexandratos, J, Wlodawer, A. | | Deposit date: | 1995-11-29 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations.
Structure, 4, 1996
|
|
3PDS
 
 | | Irreversible Agonist-Beta2 Adrenoceptor Complex | | Descriptor: | 8-hydroxy-5-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(3-sulfanylpropoxy)phenyl]ethyl}amino)ethyl]quinolin-2(1H)-one, CHOLESTEROL, Fusion protein Beta-2 adrenergic receptor/Lysozyme, ... | | Authors: | Rosenbaum, D.M, Zhang, C, Lyons, J.A, Holl, R, Aragao, D, Arlow, D.H, Rasmussen, S.G.F, Choi, H.-J, DeVree, B.T, Sunahara, R.K, Chae, P.S, Gellman, S.H, Dror, R.O, Shaw, D.E, Weis, W.I, Caffrey, M, Gmeiner, P, Kobilka, B.K. | | Deposit date: | 2010-10-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure and function of an irreversible agonist-beta(2) adrenoceptor complex
Nature, 469, 2011
|
|
2Q4D
 
 | | Ensemble refinement of the crystal structure of a lysine decarboxylase-like protein from Arabidopsis thaliana gene At5g11950 | | Descriptor: | 1,2-ETHANEDIOL, Lysine decarboxylase-like protein At5g11950, NITRATE ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
4B3L
 
 | | Family 1 6-phospho-beta-D glycosidase from Streptococcus pyogenes | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Stepper, J, Dabin, J, Ekloef, J.M, Thongpoo, P, Kongsaeree, P.T, Taylor, E.J, Turkenburg, J.P, Brumer, H, Davies, G.J. | | Deposit date: | 2012-07-24 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure and Activity of the Streptococcus Pyogenes Family Gh1 6-Phospho Beta-Glycosidase Spy1599
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5QCR
 
 | | Crystal structure of BACE complex with BMC026 | | Descriptor: | 2-(butylamino)-N-[(2S,3S,5R)-6-(butylamino)-3-hydroxy-5-methyl-6-oxo-1-phenylhexan-2-yl]-6-methoxypyridine-4-carboxamide, Beta-secretase 1, SULFATE ION | | Authors: | Rondeau, J.M, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-12-01 | | Release date: | 2020-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R grand challenge 4: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J.Comput.Aided Mol.Des., 34, 2020
|
|
3PC0
 
 | | Crystal structure of the mutant V155S of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with 6-azauridine 5'-monophosphate | | Descriptor: | 6-AZA URIDINE 5'-MONOPHOSPHATE, GLYCEROL, Orotidine 5'-monophosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Iiams, V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-10-21 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Mechanism of the orotidine 5'-monophosphate decarboxylase-catalyzed reaction: importance of residues in the orotate binding site.
Biochemistry, 50, 2011
|
|
2C8Z
 
 | | thrombin inhibitors | | Descriptor: | 1-(3-CHLOROPHENYL)METHANAMINE, DIMETHYL SULFOXIDE, HIRUDIN VARIANT-2, ... | | Authors: | Howard, N, Abell, C, Blakemore, W, Carr, R, Chessari, G, Congreve, M, Howard, S, Jhoti, H, Murray, C.W, Seavers, L.C.A, van Montfort, R.L.M. | | Deposit date: | 2005-12-08 | | Release date: | 2006-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Application of Fragment Screening and Fragment Linking to the Discovery of Novel Thrombin Inhibitors
J.Med.Chem., 49, 2006
|
|
2CCJ
 
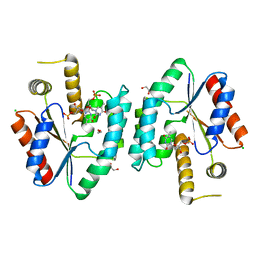 | | Crystal structure of S. aureus thymidylate kinase complexed with thymidine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
5QDR
 
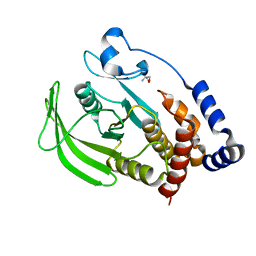 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000089a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1, methyl 3-(methylsulfonylamino)benzoate | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.784 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5QE8
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOPL000127a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(4-methoxyphenyl)-N'-pyridin-4-ylurea, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1GZR
 
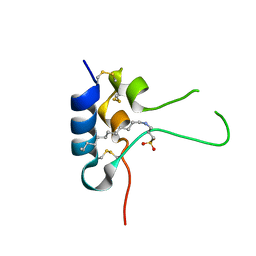 | | Human Insulin-like growth factor; ESRF data | | Descriptor: | INSULIN-LIKE GROWTH FACTOR I, N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE | | Authors: | Brzozowski, A.M, Dodson, E.J, Dodson, G.G, Murshudov, G, Verma, C, Turkenburg, J.P, De Bree, F.M, Dauter, Z. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origins of the Functional Divergence of Human Insulin-Like Growth Factor-I and Insulin
Biochemistry, 41, 2002
|
|
5QBU
 
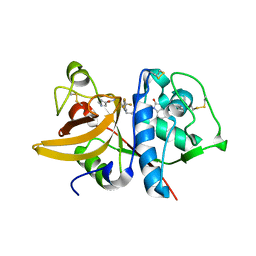 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 1-[1-(3-{5-(1H-imidazole-5-carbonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl)piperidin-4-yl]-3-methyl-1,3-dihydro-2H-benzimidazol-2-one, Cathepsin S | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5D5B
 
 | | In meso X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Liu, X, Kobilka, B, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5TN7
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-3'-fluoro-4'-hydroxy-3-((hydroxyiminio)methyl)-[1,1'-biphenyl]-4-olate | | Descriptor: | 3-fluoro-3'-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-4,4'-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Minutolo, F, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.238 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5QFO
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_XST00000644b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-fluoro-4-hydroxybenzonitrile, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
5D6L
 
 | | beta2AR-T4L - CIM | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Ma, P, Caffrey, M. | | Deposit date: | 2015-08-12 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The cubicon method for concentrating membrane proteins in the cubic mesophase.
Nat Protoc, 12, 2017
|
|
