7GAB
 
 | | PanDDA analysis group deposition -- Crystal Structure of MAP1LC3B in complex with Z1255402624 | | Descriptor: | 1,2-ETHANEDIOL, 2-tert-butylbenzene-1,4-diol, CHLORIDE ION, ... | | Authors: | Kumar, A, Marples, P.G, Tomlinson, C.W.E, Fearon, D, von-Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-08-10 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1K9E
 
 | | Crystal structure of a mutated family-67 alpha-D-glucuronidase (E285N) from Bacillus stearothermophilus T-6, complexed with 4-O-methyl-glucuronic acid | | Descriptor: | 4-O-methyl-alpha-D-glucopyranuronic acid, GLYCEROL, alpha-D-glucuronidase | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2001-10-29 | | Release date: | 2002-10-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
3NR9
 
 | | Structure of human CDC2-like kinase 2 (CLK2) | | Descriptor: | (5Z)-5-(quinolin-6-ylmethylidene)-2-[(thiophen-2-ylmethyl)amino]-1,3-thiazol-4(5H)-one, Dual specificity protein kinase CLK2 | | Authors: | Chaikuad, A, Savitsky, P, Krojer, T, Muniz, J.R.C, Filippakopoulos, P, Rellos, P, Keates, T, Fedorov, O, Pike, A.C.W, Eswaran, J, Berridge, G, Phillips, C, Zhang, Y, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of human CDC2-like kinase 2 (CLK2)
To be Published
|
|
1YRR
 
 | | Crystal Structure Of The N-Acetylglucosamine-6-Phosphate Deacetylase From Escherichia Coli K12 at 2.0 A Resolution | | Descriptor: | GLYCEROL, N-acetylglucosamine-6-phosphate deacetylase, PHOSPHATE ION | | Authors: | Ferreira, F.M, Aparicio, R, Mendoza-Hernandez, G, Calcagno, M.L, Oliva, G. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli.
J.Mol.Biol., 359, 2006
|
|
4BQ3
 
 | | Structural analysis of an exo-beta-agarase | | Descriptor: | B-AGARASE, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
1XFT
 
 | |
3RVU
 
 | | Structure of 4C1 Fab in C2221 space group | | Descriptor: | 4C1 Fab - heavy chain, 4C1 Fab - light chain | | Authors: | Chruszcz, M, Vailes, L.D, Chapman, M.D, Pomes, A, Minor, W. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular determinants for antibody binding on group 1 house dust mite allergens.
J.Biol.Chem., 287, 2012
|
|
4GKM
 
 | | Bianthranilate-like analogue bound in the outer site of anthranilate phosphoribosyltransferase (AnPRT; trpD) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-[(2-carboxyphenyl)amino]-5-methylbenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Evans, G.L, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2012-08-13 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6683 Å) | | Cite: | Repurposing the Chemical Scaffold of the Anti-Arthritic Drug Lobenzarit to Target Tryptophan Biosynthesis in Mycobacterium tuberculosis.
Chembiochem, 15, 2014
|
|
1YJC
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 50% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
1YJA
 
 | | SUBTILISIN BPN' 8397+1 (E.C. 3.4.21.14) (MUTANT WITH MET 50 REPLACED BY PHE, ASN 76 REPLACED BY ASP, GLY 169 REPLACED BY ALA, GLN 206 REPLACED BY CYS, ASN 218 REPLACED BY SER AND LYS 256 REPLACED BY TYR) (M50F, N76D, G169A, Q206C, N218S, AND K256Y) IN 20% DIMETHYLFORMAMIDE | | Descriptor: | CALCIUM ION, SUBTILISIN 8397+1 | | Authors: | Kidd, R.D, Farber, G.K. | | Deposit date: | 1996-01-16 | | Release date: | 1996-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Breaking the low barrier hydrogen bond in a serine protease.
Protein Sci., 8, 1999
|
|
4BSN
 
 | | Crystal structure of the Nuclear Export Receptor CRM1 (exportin-1) lacking the C-terminal helical extension at 4.1A | | Descriptor: | EXPORTIN-1 | | Authors: | Dian, C, Bernaudat, F, Langer, K, Oliva, M.F, Fornerod, M, Schoehn, G, Muller, C.W, Petosa, C. | | Deposit date: | 2013-06-11 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure of a Truncation Mutant of the Nuclear Export Factor Crm1 Provides Insights Into the Auto-Inhibitory Role of its C-Terminal Helix.
Structure, 21, 2013
|
|
3BZ0
 
 | | Lactobacillus Casei Thymidylate Synthase Ternary Complex with DUMP and the Phtalimidic Derivative C00 | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-(4-hydroxybiphenyl-3-yl)-4-methyl-1H-isoindole-1,3(2H)-dione, Thymidylate synthase | | Authors: | Leone, R, Cancian, L, Costi, M.P, Ferrari, S, Luciani, R, Mangani, S. | | Deposit date: | 2008-01-17 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of the binding modes of N-phenylphthalimides inhibiting bacterial thymidylate synthase through X-ray crystallography screening
J.Med.Chem., 54, 2011
|
|
2QIE
 
 | | Staphylococcus aureus molybdopterin synthase in complex with precursor Z | | Descriptor: | (2R,4AR,5AR,11AR,12AS)-8-AMINO-2-HYDROXY-4A,5A,9,11,11A,12A-HEXAHYDRO[1,3,2]DIOXAPHOSPHININO[4',5':5,6]PYRANO[3,2-G]PTERIDINE-10,12(4H,6H)-DIONE 2-OXIDE, Molybdopterin synthase small subunit, Molybdopterin-converting factor subunit 2 | | Authors: | Daniels, J.N, Schindelin, H. | | Deposit date: | 2007-07-04 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a molybdopterin synthase-precursor Z complex: insight into its sulfur transfer mechanism and its role in molybdenum cofactor deficiency.
Biochemistry, 47, 2008
|
|
3SBK
 
 | | Russell's viper venom serine proteinase, RVV-V (PPACK-bound form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Vipera russelli proteinase RVV-V gamma | | Authors: | Nakayama, D, Ben Ammar, Y, Takeda, S. | | Deposit date: | 2011-06-05 | | Release date: | 2011-09-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of coagulation factor V recognition for cleavage by RVV-V
Febs Lett., 585, 2011
|
|
2QX1
 
 | | Crystal structure of the complex between mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FABH) and decyl-COA disulfide | | Descriptor: | Beta-ketoacyl-ACP synthase III, COENZYME A, DECANE-1-THIOL | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-08-10 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing reactivity and substrate specificity of both subunits of the dimeric Mycobacterium tuberculosis FabH using alkyl-CoA disulfide inhibitors and acyl-CoA substrates
Bioorg.Chem., 36, 2008
|
|
3SDZ
 
 | | Structural characterization of the subunit A mutant F427W of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Tadwal, V.S, Manimekalai, M.S.S, Balakrishna, A.M, Gruber, G. | | Deposit date: | 2011-06-09 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Engineered tryptophan in the adenine-binding pocket of catalytic subunit A of A-ATP synthase demonstrates the importance of aromatic residues in adenine binding, forming a tool for steady-state and time-resolved fluorescence spectroscopy.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
6M9T
 
 | | Crystal structure of EP3 receptor bound to misoprostol-FA | | Descriptor: | (11alpha,12alpha,13E,16S)-11,16-dihydroxy-16-methyl-9-oxoprost-13-en-1-oic acid, (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, ... | | Authors: | Audet, M, White, K.L, Breton, B, Zarzycka, B, Han, G.W, Lu, Y, Gati, C, Batyuk, A, Popov, P, Velasquez, J, Manahan, D, Hu, H, Weierstall, U, Liu, W, Shui, W, Katrich, V, Cherezov, V, Hanson, M.A, Stevens, R.C. | | Deposit date: | 2018-08-24 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of misoprostol bound to the labor inducer prostaglandin E2receptor.
Nat. Chem. Biol., 15, 2019
|
|
4KNH
 
 | | Structure of the Chaetomium thermophilum adaptor nucleoporin Nup192 N-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Nup192p, ... | | Authors: | Stuwe, T, Lin, D.H, Collins, L.N, Hoelz, A. | | Deposit date: | 2013-05-09 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2D4N
 
 | | Crystal Structure of M-PMV dUTPase complexed with dUPNPP, substrate analogue | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DU, ... | | Authors: | Nemeth, V, Barabas, O, Vertessy, G.B. | | Deposit date: | 2005-10-20 | | Release date: | 2006-11-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins.
Nucleic Acids Res., 35, 2007
|
|
3RZ4
 
 | |
3RVY
 
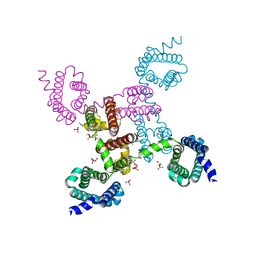 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.7 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3O6L
 
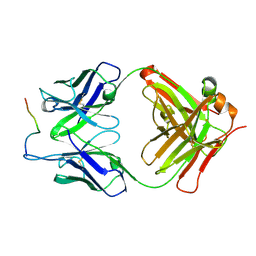 | |
4C70
 
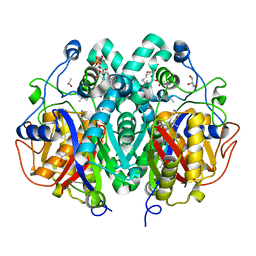 | | Crystal structure of M. tuberculosis C171Q KasA in complex with TLM4 | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(10R)-10-methyloctadecanoyl]oxy}propyl phosphate, (5R)-4-hydroxy-5-methyl-5-[(1E)-2-methylbuta-1,3-dien-1-yl]-3-propylthiophen-2(5H)-one, 3-OXOACYL-[ACYL-CARRIER-PROTEIN] SYNTHASE 1, ... | | Authors: | Schiebel, J, Kapilashrami, K, Fekete, A, Bommineni, G.R, Schaefer, C.M, Mueller, M.J, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-09-19 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for the Recognition of Mycolic Acid Precursors by Kasa, a Condensing Enzyme and Drug Target from Mycobacterium Tuberculosis
J.Biol.Chem., 288, 2013
|
|
4CAV
 
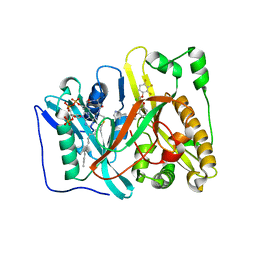 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a benzofuran ligand R0-09-4879 | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, CHLORIDE ION, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Fang, W, Raimi, O.G, Blair, D.E, Harrison, J, Ruda, G.F, Lockhart, D.E.A, Torrie, L.S, Wyatt, P.G, Gilbert, I.H, Van Aalten, D.M.F. | | Deposit date: | 2013-10-09 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | N-Myristoyltransferase is a Cell Wall Target in Aspergillus Fumigatus.
Acs Chem.Biol., 10, 2015
|
|
4GK2
 
 | | Human EphA3 Kinase domain in complex with ligand 66 | | Descriptor: | 7-(5-hydroxy-2-methylphenyl)-8-(2-methoxyphenyl)-1-methyl-1H-imidazo[2,1-f]purine-2,4(3H,8H)-dione, EPH receptor A3 | | Authors: | Dong, J, Caflisch, A. | | Deposit date: | 2012-08-10 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Optimization of Inhibitors of the Tyrosine Kinase EphB4. 2. Cellular Potency Improvement and Binding Mode Validation by X-ray Crystallography.
J.Med.Chem., 56, 2013
|
|
