4LQW
 
 | |
7JS3
 
 | | Structure of the Class II Fructose-1,6-Bisphophatase from Francisella tularensis | | Descriptor: | Fructose-1,6-bisphosphatase, GLYCEROL, MAGNESIUM ION | | Authors: | Abad-Zapatero, C, Wolf, N.M, Movahedzadeh, F, Gutka, H, Selezneva, A. | | Deposit date: | 2020-08-13 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of the class II fructose-1,6-bisphosphatase from Francisella tularensis.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2BSZ
 
 | | Structure of Mesorhizobium loti arylamine N-acetyltransferase 1 | | Descriptor: | ARYLAMINE N-ACETYLTRANSFERASE 1 | | Authors: | Holton, S.J, Dairou, J, Sandy, J, Rodrigues-Lima, F, Dupret, J.-M, Noble, M.E.M, Sim, E. | | Deposit date: | 2005-05-24 | | Release date: | 2005-05-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Mesorhizobium Loti Arylamine N-Acetyltransferase 1.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
3OS7
 
 | |
3VW6
 
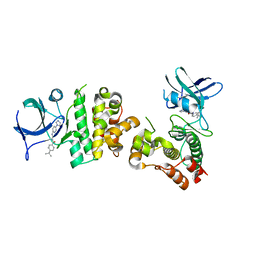 | | Crystal structure of human apoptosis signal-regulating kinase 1 (ASK1) with imidazopyridine inhibitor | | Descriptor: | 4-tert-butyl-N-[6-(1H-imidazol-1-yl)imidazo[1,2-a]pyridin-2-yl]benzamide, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Terao, Y, Suzuki, H, Yoshikawa, M, Yashiro, H, Takekawa, S, Fujitani, Y, Okada, K, Inoue, Y, Yamamoto, Y, Nakagawa, H, Yao, S, Kawamoto, T, Uchikawa, O. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and biological evaluation of imidazo[1,2-a]pyridines as novel and potent ASK1 inhibitors.
Bioorg. Med. Chem. Lett., 22, 2012
|
|
4MJF
 
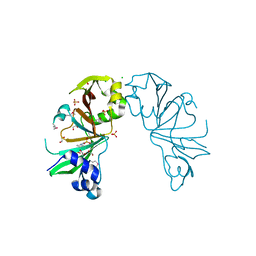 | |
4R4H
 
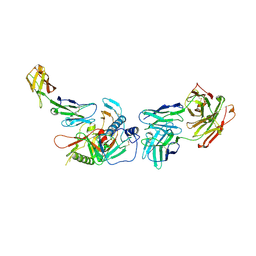 | | Crystal structure of non-neutralizing, A32-like antibody 2.2c in complex with HIV-1 Env gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2.2c, Heavy chain, ... | | Authors: | Mclellan, J, Acharya, P, Huang, C.-C, Kwong, P.D. | | Deposit date: | 2014-08-19 | | Release date: | 2014-09-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (4.28 Å) | | Cite: | Structural Definition of an Antibody-Dependent Cellular Cytotoxicity Response Implicated in Reduced Risk for HIV-1 Infection.
J.Virol., 88, 2014
|
|
3OST
 
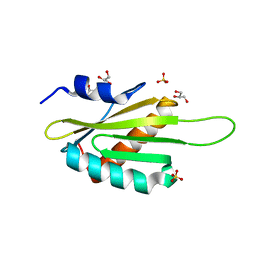 | | Structure of the Kinase Associated-1 (KA1) from Kcc4p | | Descriptor: | GLYCEROL, SULFATE ION, serine/threonine-protein kinase KCC4 | | Authors: | Moravcevic, K, Lemmon, M.A. | | Deposit date: | 2010-09-09 | | Release date: | 2010-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Kinase Associated-1 Domains Drive MARK/PAR1 Kinases to Membrane Targets by Binding Acidic Phospholipids.
Cell(Cambridge,Mass.), 143, 2010
|
|
1TE6
 
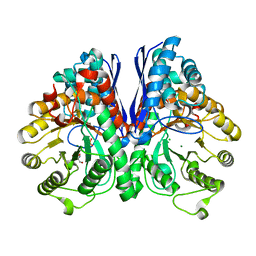 | | Crystal Structure of Human Neuron Specific Enolase at 1.8 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Gamma enolase, ... | | Authors: | Chai, G, Brewer, J, Lovelace, L, Aoki, T, Minor, W, Lebioda, L. | | Deposit date: | 2004-05-24 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Expression, Purification and the 1.8 A Resolution Crystal Structure of Human Neuron Specific Enolase
J.Mol.Biol., 341, 2004
|
|
3RJV
 
 | |
1IHW
 
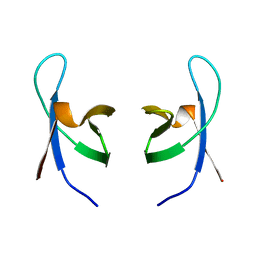 | | SOLUTION STRUCTURE OF THE DNA BINDING DOMAIN OF HIV-1 INTEGRASE, NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE | | Authors: | Clore, G.M, Lodi, P.J, Ernst, J.A, Gronenborn, A.M. | | Deposit date: | 1995-05-12 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of HIV-1 integrase.
Biochemistry, 34, 1995
|
|
1ITG
 
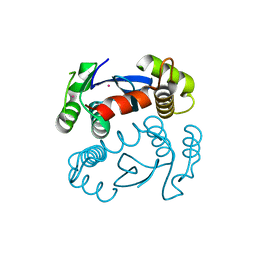 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HIV-1 INTEGRASE: SIMILARITY TO OTHER POLYNUCLEOTIDYL TRANSFERASES | | Descriptor: | CACODYLATE ION, HIV-1 INTEGRASE | | Authors: | Dyda, F, Hickman, A.B, Jenkins, T.M, Engelman, A, Craigie, R, Davies, D.R. | | Deposit date: | 1994-11-21 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases.
Science, 266, 1994
|
|
3H0S
 
 | |
3TC8
 
 | |
1CRM
 
 | | STRUCTURE AND FUNCTION OF CARBONIC ANHYDRASES | | Descriptor: | CARBONIC ANHYDRASE I, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Yadava, V.S, Kannan, K.K. | | Deposit date: | 1994-03-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Carbonic Anhydrases
Biomolecular Structure, Conformation, Function and Evolution, 1, 1981
|
|
4O0R
 
 | |
1LLF
 
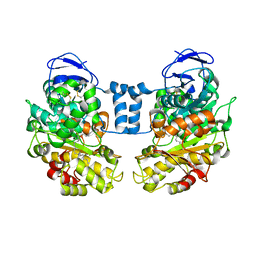 | | Cholesterol Esterase (Candida Cylindracea) Crystal Structure at 1.4A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase 3, TRICOSANOIC ACID | | Authors: | Pletnev, V, Addlagatta, A, Wawrzak, Z, Duax, W. | | Deposit date: | 2002-04-28 | | Release date: | 2003-01-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Three-dimensional structure of homodimeric cholesterol esterase-ligand complex at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1LRA
 
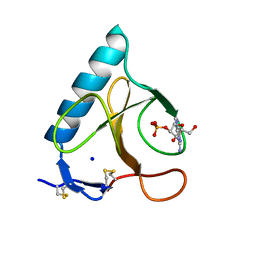 | | CRYSTALLOGRAPHIC STUDY OF GLU 58 ALA RNASE T1(ASTERISK)2'-GUANOSINE MONOPHOSPHATE AT 1.9 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE T1, SODIUM ION | | Authors: | Pletinckx, J, Steyaert, J, Choe, H.-W, Heinemann, U, Wyns, L. | | Deposit date: | 1993-10-01 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of Glu58Ala RNase T1 x 2'-guanosine monophosphate at 1.9-A resolution.
Biochemistry, 33, 1994
|
|
1YND
 
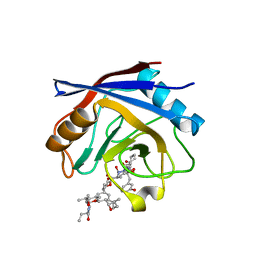 | | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6A resolution | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, SANGLIFEHRIN A | | Authors: | Kallen, J, Sedrani, R, Zenke, G, Wagner, J. | | Deposit date: | 2005-01-24 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human cyclophilin A in complex with the novel immunosuppressant sanglifehrin A at 1.6 A resolution.
J.Biol.Chem., 280, 2005
|
|
4NKK
 
 | |
4J1W
 
 | |
4EP3
 
 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate CA-p2 | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-16 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
3EDO
 
 | |
3TTQ
 
 | | Crystal structure of Leuconostoc mesenteroides NRRL B-1299 N-terminally truncated dextransucrase DSR-E in orthorhombic apo-form at 1.9 angstrom resolution | | Descriptor: | CALCIUM ION, Dextransucrase, GLYCEROL, ... | | Authors: | Brison, Y, Pijning, T, Fabre, E, Mourey, L, Morel, S, Potocki-Veronese, G, Monsan, P, Remaud-Simeon, M, Dijkstra, B.W, Tranier, S. | | Deposit date: | 2011-09-15 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and structural characterization of alpha-(1-2) branching sucrase derived from DSR-E glucansucrase
J.Biol.Chem., 287, 2012
|
|
4ISL
 
 | | Crystal Structure of the inactive Matriptase in complex with its inhibitor HAI-1 | | Descriptor: | GLUTATHIONE, GLYCEROL, Kunitz-type protease inhibitor 1, ... | | Authors: | Huang, M.D, Zhao, B.Y, Yuan, C, Li, R. | | Deposit date: | 2013-01-16 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structures of matriptase in complex with its inhibitor hepatocyte growth factor activator inhibitor-1.
J.Biol.Chem., 288, 2013
|
|
