8IUG
 
 | | Cryo-EM structure of the RC-LH core complex from roseiflexus castenholzii | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-03-24 | | Release date: | 2023-11-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
2P84
 
 | | Crystal structure of ORF041 from Bacteriophage 37 | | Descriptor: | ORF041 | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Bain, K.T, Adams, J.M, Reyes, C, Lau, C, Gilmore, J, Rooney, I, Wasserman, T, Gheyi, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-21 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the hypothetical protein from Staphylococcus phage 37
To be Published
|
|
2P8J
 
 | |
2P91
 
 | | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5 | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Chen, L, Li, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Enoyl-[acyl-carrier-protein] reductase (NADH) from Aquifex aeolicus VF5
To be Published
|
|
2P9N
 
 | |
2PA1
 
 | | Structure of the PDZ domain of human PDLIM2 bound to a C-terminal extension from human beta-tropomyosin | | Descriptor: | CHLORIDE ION, PDZ and LIM domain protein 2 | | Authors: | Uppenberg, J, Shrestha, L, Elkins, J, Burgess-Brown, N, Salah, E, Bunkoczi, G, Papagrigoriou, E, Pike, A.C.W, Turnbull, A.P, Ugochukwu, E, Umeano, C, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-27 | | Release date: | 2007-05-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms
Protein Sci., 19, 2010
|
|
2PCE
 
 | | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM | | Descriptor: | PHOSPHATE ION, putative mandelate racemase/muconate lactonizing enzyme | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Lau, C, Ozyurt, S, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of putative mandelate racemase/muconate lactonizing enzyme from Roseovarius nubinhibens ISM
To be Published
|
|
2PBY
 
 | | Probable Glutaminase from Geobacillus kaustophilus HTA426 | | Descriptor: | Glutaminase | | Authors: | Dillard, B.D, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Rose, J.P, Wang, B.-C, RIKEN Structural Genomics/Proteomics Initiative (RSGI), Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-03-29 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Glutaminase from Geobacillus kaustophilus HTA426
To be Published
|
|
1M04
 
 | | Mutant Streptomyces plicatus beta-hexosaminidase (D313N) in complex with product (GlcNAc) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, CHLORIDE ION, ... | | Authors: | Williams, S.J, Mark, B.L, Vocadlo, D.J, James, M.N.G, Withers, S.G. | | Deposit date: | 2002-06-11 | | Release date: | 2002-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical
role in substrate-assisted catalysis by orienting the 2-acetamido group
and stabilizing the transition state.
J.Biol.Chem., 277, 2002
|
|
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
8IUN
 
 | | Cryo-EM structure of the CRT-LESS RC-LH core complex from roseiflexus castenholzii | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-03-24 | | Release date: | 2023-12-06 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | New insights on the photocomplex of Roseiflexus castenholzii revealed from comparisons of native and carotenoid-depleted complexes.
J.Biol.Chem., 299, 2023
|
|
2PEN
 
 | | Crystal structure of RbcX, crystal form I | | Descriptor: | ORF134 | | Authors: | Saschenbrecker, S, Bracher, A, Vasudeva Rao, K, Vasudeva Rao, B, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2007-04-03 | | Release date: | 2007-07-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of RbcX, an Assembly Chaperone for Hexadecameric Rubisco.
Cell(Cambridge,Mass.), 129, 2007
|
|
2PF4
 
 | |
5EZO
 
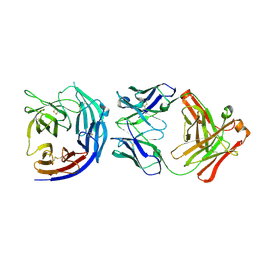 | | Crystal Structure of PfCyRPA in complex with an invasion-inhibitory antibody Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PfCyRPA, c12 FAB, ... | | Authors: | Favuzza, P, Pluschke, G, Rudolph, M.G. | | Deposit date: | 2015-11-26 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structure of the malaria vaccine candidate antigen CyRPA and its complex with a parasite invasion inhibitory antibody.
Elife, 6, 2017
|
|
2OL9
 
 | |
2ONV
 
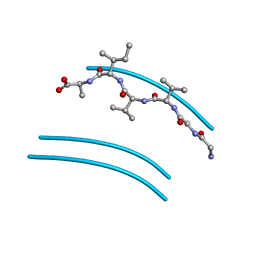 | |
2OMP
 
 | |
2OMZ
 
 | |
2ONZ
 
 | | Structure of K57A hPNMT with inhibitor 7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | N-(4-CHLOROPHENYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase
J.Med.Chem., 50, 2007
|
|
2OOR
 
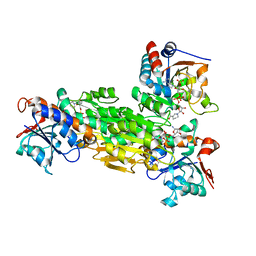 | | Structure of transhydrogenase (dI.NAD+)2(dIII.H2NADPH)1 asymmetric complex | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Bhakta, T, Jackson, J.B. | | Deposit date: | 2007-01-26 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Biochemistry, 46, 2007
|
|
2ONK
 
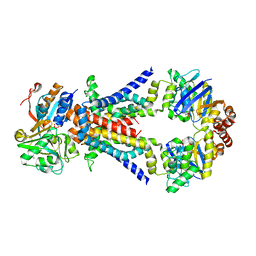 | | ABC transporter ModBC in complex with its binding protein ModA | | Descriptor: | MAGNESIUM ION, Molybdate/tungstate ABC transporter, ATP-binding protein, ... | | Authors: | Hollenstein, K, Frei, D.C, Locher, K.P. | | Deposit date: | 2007-01-24 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of an ABC transporter in complex with its binding protein.
Nature, 446, 2007
|
|
2OPB
 
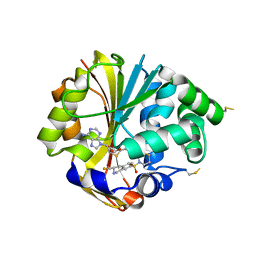 | | Structure of K57A hPNMT with inhibitor 3-fluoromethyl-7-thiomorpholinosulfonamide-THIQ and AdoHcy | | Descriptor: | (3R)-3-(FLUOROMETHYL)-7-(THIOMORPHOLIN-4-YLSULFONYL)-1,2,3,4-TETRAHYDROISOQUINOLINE, PHOSPHATE ION, Phenylethanolamine N-methyltransferase, ... | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2007-01-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Enzyme Adaptation to Inhibitor Binding: A Cryptic Binding Site in Phenylethanolamine N-Methyltransferase.
J.Med.Chem., 50, 2007
|
|
2OQR
 
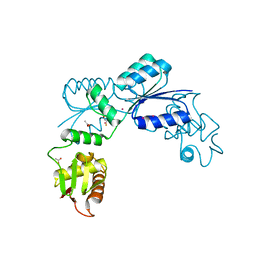 | |
2OT4
 
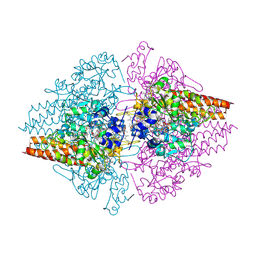 | | Structure of a hexameric multiheme c nitrite reductase from the extremophile bacterium Thiolkalivibrio nitratireducens | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | Polyakov, K.M, Boyko, K.M, Slutsky, A, Tikhonova, T.V, Antipov, A.N, Zvyagilskaya, R.A, Popov, A.N, Lamzin, V.S, Bourenkov, G.P, Popov, V.O. | | Deposit date: | 2007-02-07 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution structural analysis of a novel octaheme cytochrome c nitrite reductase from the haloalkaliphilic bacterium Thioalkalivibrio nitratireducens.
J.Mol.Biol., 389, 2009
|
|
2OQX
 
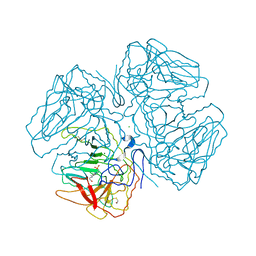 | | Crystal Structure of the apo form of E. coli tryptophanase at 1.9 A resolution | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Goldgur, Y, Kogan, A, Gdalevsky, G, Parola, A, Cohen-Luria, R, Almog, O. | | Deposit date: | 2007-02-01 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of apo tryptophanase from Escherichia coli reveals a wide-open conformation.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
