6GSJ
 
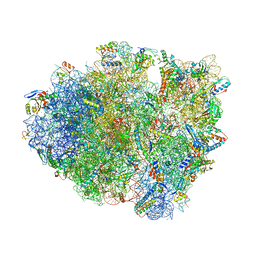 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAThr in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.
Nucleic Acids Res., 46, 2018
|
|
6GSK
 
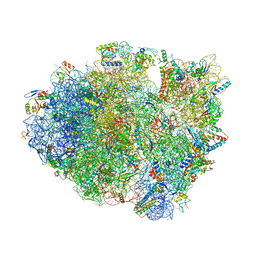 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and near-cognate tRNAThr in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.
Nucleic Acids Res., 46, 2018
|
|
6GSL
 
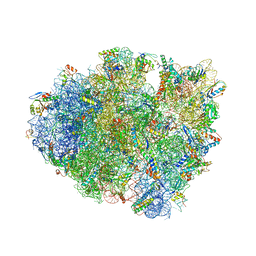 | | Structure of T. thermophilus 70S ribosome complex with mRNA, tRNAfMet and cognate tRNAArg in the A-site | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2018-06-14 | | Release date: | 2018-07-04 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Tautomeric G•U pairs within the molecular ribosomal grip and fidelity of decoding in bacteria.
Nucleic Acids Res., 46, 2018
|
|
6GSM
 
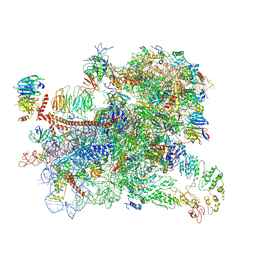 | | Structure of a partial yeast 48S preinitiation complex in open conformation. | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S0, 40S ribosomal protein S1, ... | | Authors: | Llacer, J.L, Hussain, T, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2018-06-14 | | Release date: | 2019-07-31 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Large-scale movement of eIF3 domains during translation initiation modulate start codon selection.
Nucleic Acids Res., 2021
|
|
6GSN
 
 | |
6GWT
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-25 | | Release date: | 2018-08-15 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GXM
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GXN
 
 | | Cryo-EM structure of an E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and Pint-tRNA (State III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GXO
 
 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP, RF1(GAQ) and P/E-tRNA (State IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GXP
 
 | | Cryo-EM structure of a rotated E. coli 70S ribosome in complex with RF3-GDPCP(RF3-only) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Huter, P, Maracci, C, Peterek, M, Rodnina, M.V, Wilson, D.N. | | Deposit date: | 2018-06-27 | | Release date: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Visualization of translation termination intermediates trapped by the Apidaecin 137 peptide during RF3-mediated recycling of RF1.
Nat Commun, 9, 2018
|
|
6GZQ
 
 | | T. thermophilus hibernating 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Flygaard, R.K, Jenner, L.B. | | Deposit date: | 2018-07-04 | | Release date: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism.
Nat Commun, 9, 2018
|
|
6GZX
 
 | | T. thermophilus hibernating 100S ribosome (ice) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Flygaard, R.K, Jenner, L.B. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (4.57 Å) | | Cite: | Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism.
Nat Commun, 9, 2018
|
|
6GZZ
 
 | | T. thermophilus hibernating 100S ribosome (amc) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Flygaard, R.K, Jenner, L.B. | | Deposit date: | 2018-07-05 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.13 Å) | | Cite: | Cryo-EM structure of the hibernating Thermus thermophilus 100S ribosome reveals a protein-mediated dimerization mechanism.
Nat Commun, 9, 2018
|
|
6H4N
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - 70S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, N.D. | | Deposit date: | 2018-07-22 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6H58
 
 | | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1 - Full 100S Hibernating E. coli Ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Beckert, B, Turk, M, Czech, A, Berninghausen, O, Beckmann, R, Ignatova, Z, Plitzko, J, Wilson, D.N. | | Deposit date: | 2018-07-24 | | Release date: | 2018-09-05 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.
Nat Microbiol, 3, 2018
|
|
6HA1
 
 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HA8
 
 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCJ
 
 | | Structure of the rabbit 80S ribosome on globin mRNA in the rotated state with A/P and P/E tRNAs | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCQ
 
 | | Structure of the rabbit collided di-ribosome (collided monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-16 | | Release date: | 2018-10-17 | | Last modified: | 2018-11-14 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HHQ
 
 | | Crystal structure of compound C45 bound to the yeast 80S ribosome | | Descriptor: | (3~{R})-3-[(1~{S})-2-[(1~{S},4~{a}~{R},6~{S},7~{S},8~{a}~{R})-6,7-bis(chloranyl)-5,5,8~{a}-trimethyl-2-methylidene-3,4,4~{a},6,7,8-hexahydro-1~{H}-naphthalen-1-yl]-1-oxidanyl-ethyl]pyrrolidine-2,5-dione, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Pellegrino, S, Vanderwal, C.D, Yusupov, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.10000038 Å) | | Cite: | Understanding the role of intermolecular interactions between lissoclimides and the eukaryotic ribosome.
Nucleic Acids Res., 47, 2019
|
|
6HRM
 
 | | E. coli 70S d2d8 stapled ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schmied, W.H, Tnimov, Z, Uttamapinant, C, Rae, C.D, Fried, S.D, Chin, J.W. | | Deposit date: | 2018-09-27 | | Release date: | 2018-12-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Controlling orthogonal ribosome subunit interactions enables evolution of new function.
Nature, 564, 2018
|
|
6HTQ
 
 | |
6I7O
 
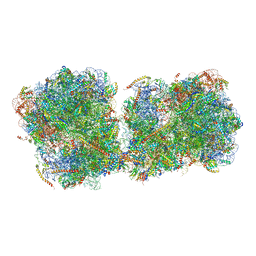 | | The structure of a di-ribosome (disome) as a unit for RQC and NGD quality control pathways recognition. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2018-11-16 | | Release date: | 2019-01-16 | | Last modified: | 2019-03-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways.
EMBO J., 38, 2019
|
|
