2KUM
 
 | | Solution structure of the human chemokine CCL27 | | Descriptor: | C-C motif chemokine 27 | | Authors: | Kirkpatrick, J.P, Jansma, A, Hsu, A, Handel, T.M, Nietlispach, D. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR analysis of the structure, dynamics, and unique oligomerization properties of the chemokine CCL27.
J.Biol.Chem., 285, 2010
|
|
2AFF
 
 | | The solution structure of the Ki67FHA/hNIFK(226-269)3P complex | | Descriptor: | Antigen KI-67, MKI67 FHA domain interacting nucleolar phosphoprotein | | Authors: | Byeon, I.-J.L, Li, H, Song, H, Gronenborn, A.M, Tsai, M.D. | | Deposit date: | 2005-07-25 | | Release date: | 2005-10-25 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Sequential phosphorylation and multisite interactions characterize specific target recognition by the FHA domain of Ki67.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2N6Y
 
 | |
2N6Z
 
 | |
1TL1
 
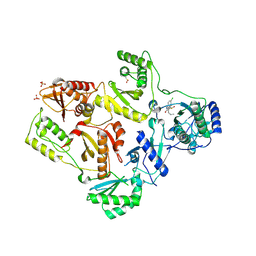 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW451211 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLSULFINYL)-3-PROPYLQUINOLIN-2(1H)-ONE, PHOSPHATE ION, Pol polyprotein, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
5KVS
 
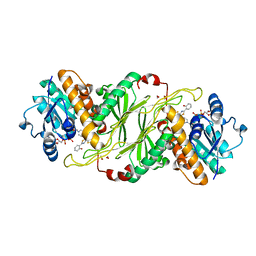 | | Substrate Analog and NADP+ bound structure of Irp3, a Thiazolinyl Imine Reductase from Yersinia enterocolitica | | Descriptor: | (4~{R})-2-[2-(2-hydroxyphenyl)-1,3-thiazol-4-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Meneely, K.M, Lamb, A.L. | | Deposit date: | 2016-07-15 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Holo Structure and Steady State Kinetics of the Thiazolinyl Imine Reductases for Siderophore Biosynthesis.
Biochemistry, 55, 2016
|
|
3LOG
 
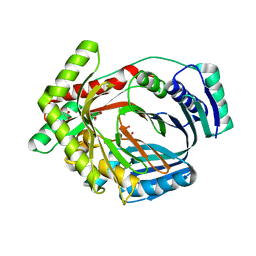 | | Crystal structure of MbtI from Mycobacterium tuberculosis | | Descriptor: | CARBONATE ION, GLYCEROL, Isochorismate synthase/isochorismate-pyruvate lyase mbtI, ... | | Authors: | Bulloch, E.M.M, Lott, J.S, Baker, E.N, Johnston, J.M. | | Deposit date: | 2010-02-03 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Inhibition studies of Mycobacterium tuberculosis salicylate synthase (MbtI).
Chemmedchem, 5, 2010
|
|
2BRA
 
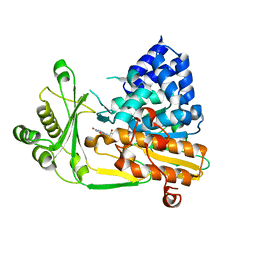 | | Structure of N-Terminal FAD Binding motif of mouse MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9 INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Nadella, M, Bianchet, M.A, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2005-05-04 | | Release date: | 2005-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and activity of the axon guidance protein MICAL.
Proc. Natl. Acad. Sci. U.S.A., 102, 2005
|
|
1IH7
 
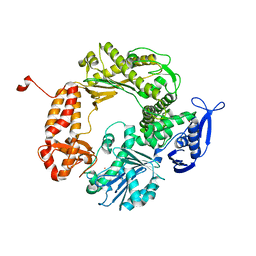 | | High-Resolution Structure of Apo RB69 DNA Polymerase | | Descriptor: | DNA POLYMERASE, GUANOSINE, POTASSIUM ION | | Authors: | Franklin, M.C, Wang, J, Steitz, T.A. | | Deposit date: | 2001-04-18 | | Release date: | 2001-06-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure of the Replicating Complex of a Pol alpha Family DNA Polymerase
Cell(Cambridge,Mass.), 105, 2001
|
|
1RTJ
 
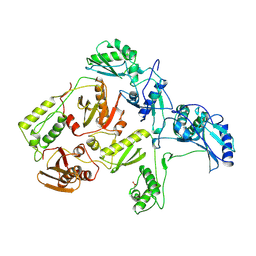 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
5KVQ
 
 | |
5LFD
 
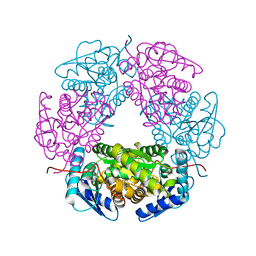 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ramazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-01 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
1S1T
 
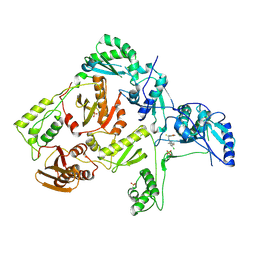 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with UC-781 | | Descriptor: | 2-METHYL-FURAN-3-CARBOTHIOIC ACID [4-CHLORO-3-(3-METHYL-BUT-2-ENYLOXY)-PHENYL]-AMIDE, PHOSPHATE ION, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1EXM
 
 | | CRYSTAL STRUCTURE OF THERMUS THERMOPHILUS ELONGATION FACTOR TU (EF-TU) IN COMPLEX WITH THE GTP ANALOGUE GPPNHP. | | Descriptor: | ELONGATION FACTOR TU (EF-TU), MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Hogg, T, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2000-05-03 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the GTPase Mechanism of EF-Tu from Structural Studies
The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions, 28, 2000
|
|
5LG5
 
 | | Crystal structure of allantoin racemase from Pseudomonas fluorescens AllR | | Descriptor: | Allantoin racemase | | Authors: | Cendron, l, Zanotti, G, Percudani, R, Ragazzina, I, Puggioni, V, Maccacaro, E, Liuzzi, A, Secchi, A. | | Deposit date: | 2016-07-06 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure and Function of a Microbial Allantoin Racemase Reveal the Origin and Conservation of a Catalytic Mechanism.
Biochemistry, 55, 2016
|
|
5LMC
 
 | | Oxidized flavodiiron core of Escherichia coli flavorubredoxin, including the Fe-4SG atoms from its rubredoxin domain | | Descriptor: | ACETIC ACID, Anaerobic nitric oxide reductase flavorubredoxin, CACODYLATE ION, ... | | Authors: | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
5EX6
 
 | |
5LLD
 
 | | Flavodiiron core of Escherichia coli flavorubredoxin in the reduced form. | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Romao, C.V, Borges, P.T, Vicente, J.B, Carrondo, M.A, Teixeira, M, Frazao, C. | | Deposit date: | 2016-07-27 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure of Escherichia coli Flavodiiron Nitric Oxide Reductase.
J.Mol.Biol., 428, 2016
|
|
1S1X
 
 | | Crystal structure of V108I mutant HIV-1 reverse transcriptase in complex with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
1FK9
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DMP-266(EFAVIRENZ) | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-09 | | Release date: | 2000-11-03 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
STRUCTURE FOLD.DES., 8, 2000
|
|
1S5Q
 
 | | Solution Structure of Mad1 SID-mSin3A PAH2 Complex | | Descriptor: | MAD protein, Sin3a protein | | Authors: | Swanson, K.A, Knoepfler, P.S, Huang, K, Kang, R.S, Cowley, S.M, Laherty, C.D, Eisenman, R.N, Radhakrishnan, I. | | Deposit date: | 2004-01-21 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3RV8
 
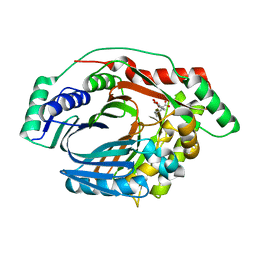 | | Structure of a M. tuberculosis Salicylate Synthase, MbtI, in Complex with an Inhibitor with Cyclopropyl R-Group | | Descriptor: | 3-{[(E)-1-carboxy-2-cyclopropylethenyl]oxy}-2-hydroxybenzoic acid, 3-{[(Z)-1-carboxy-2-cyclopropylethenyl]oxy}-2-hydroxybenzoic acid, Isochorismate synthase/isochorismate-pyruvate lyase mbtI | | Authors: | Chi, G, Bulloch, E.M.M, Manos-Turvey, A, Payne, R.J, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 51, 2012
|
|
1FKP
 
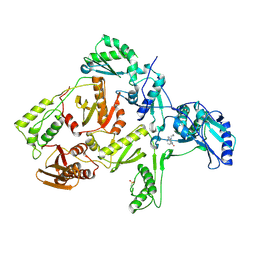 | | CRYSTAL STRUCTURE OF NNRTI RESISTANT K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Milton, J, Weaver, K.L, Short, S.A, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-08-10 | | Release date: | 2000-11-03 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the resilience of efavirenz (DMP-266) to drug resistance mutations in HIV-1 reverse transcriptase.
Structure Fold.Des., 8, 2000
|
|
1S1V
 
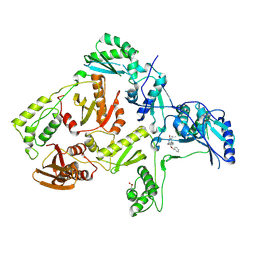 | | Crystal structure of L100I mutant HIV-1 reverse transcriptase in complex with TNK-651 | | Descriptor: | 6-BENZYL-1-BENZYLOXYMETHYL-5-ISOPROPYL URACIL, Reverse transcriptase | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2004-01-07 | | Release date: | 2004-06-29 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors
J.Mol.Biol., 336, 2004
|
|
3RV7
 
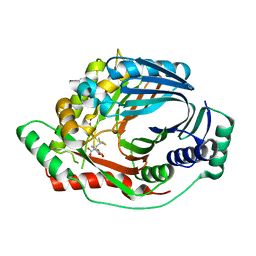 | | Structure of a M. tuberculosis Salicylate Synthase, MbtI, in Complex with an Inhibitor with Isopropyl R-Group | | Descriptor: | 3-{[(1Z)-1-carboxy-3-methylbut-1-en-1-yl]oxy}-2-hydroxybenzoic acid, Isochorismate synthase/isochorismate-pyruvate lyase mbtI | | Authors: | Chi, G, Bulloch, E.M.M, Manos-Turvey, A, Payne, R.J, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 51, 2012
|
|
