5K2M
 
 | | Bifunctional LysX/ArgX from Thermococcus kodakarensis with LysW-gamma-AAA | | Descriptor: | 2-AMINOHEXANEDIOIC ACID, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yoshida, A, Tomita, T, Nishiyama, M. | | Deposit date: | 2016-05-19 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Lysine Biosynthesis of Thermococcus kodakarensis with the Capacity to Function as an Ornithine Biosynthetic System.
J. Biol. Chem., 291, 2016
|
|
5K5U
 
 | |
4V52
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V7W
 
 | | Structure of the Thermus thermophilus ribosome complexed with chloramphenicol. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Bulkley, D.P, Innis, C.A, Blaha, G, Steitz, T.A. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Revisiting the structures of several antibiotics bound to the bacterial ribosome.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V8C
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-leu complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V4H
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with the antibiotic kasugamyin at 3.5A resolution. | | Descriptor: | (1S,2R,3S,4R,5S,6S)-2,3,4,5,6-PENTAHYDROXYCYCLOHEXYL 2-AMINO-4-{[CARBOXY(IMINO)METHYL]AMINO}-2,3,4,6-TETRADEOXY-ALPHA-D-ARABINO-HEXOPYRANOSIDE, 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, ... | | Authors: | Schuwirth, B.S, Vila-Sanjurjo, A, Cate, J.H.D. | | Deposit date: | 2006-08-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structural analysis of kasugamycin inhibition of translation.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4W2I
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with negamycin, mRNA and three deacylated tRNAs in the A, P and E sites | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Szal, T, Jiang, F, Gupta, P, Matsuda, R, Shiozuka, M, Steitz, T.A, Vazquez-Laslop, N, Mankin, A.S. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Negamycin Interferes with Decoding and Translocation by Simultaneous Interaction with rRNA and tRNA.
Mol.Cell, 56, 2014
|
|
4V57
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with spectinomycin and neomycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Shoji, S, Holton, J.M, Fredrick, K, Cate, J.H.D. | | Deposit date: | 2007-07-21 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A steric block in translation caused by the antibiotic spectinomycin.
Acs Chem.Biol., 2, 2007
|
|
4V8G
 
 | | Crystal structure of RMF bound to the 70S ribosome. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Blaha, G.M, Steitz, T.A. | | Deposit date: | 2011-12-11 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.
Science, 336, 2012
|
|
4V9D
 
 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
4W2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V9B
 
 | | Crystal Structure of the 70S ribosome with tigecycline. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2012-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4V7L
 
 | | The structures of viomycin bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-11-12 | | Release date: | 2014-07-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of the anti-tuberculosis antibiotics viomycin and capreomycin bound to the 70S ribosome.
Nat.Struct.Mol.Biol., 17, 2010
|
|
4V87
 
 | | Crystal structure analysis of ribosomal decoding. | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-09-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V5E
 
 | | Insights into translational termination from the structure of RF2 bound to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Weixlbaumer, A, Jin, H, Neubauer, C, Voorhees, R.M, Petry, S, Kelley, A.C, Ramakrishnan, V. | | Deposit date: | 2009-04-30 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Insights Into Translational Termination from the Structure of Rf2 Bound to the Ribosome.
Science, 322, 2008
|
|
2PSM
 
 | | Crystal structure of Interleukin 15 in complex with Interleukin 15 receptor alpha | | Descriptor: | BENZAMIDINE, Interleukin-15, Interleukin-15 receptor alpha chain | | Authors: | Olsen, S.K, Murayama, K, Kishishita, S, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Ota, N, Kanagawa, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the Interleukin-15{middle dot}Interleukin-15 Receptor {alpha} Complex: INSIGHTS INTO TRANS AND CIS PRESENTATION
J.Biol.Chem., 282, 2007
|
|
2QC9
 
 | |
5T0C
 
 | | Structural basis for dynamic regulation of the human 26S proteasome | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Chen, S, Wu, J, Lu, Y, Ma, Y.B, Lee, B.H, Yu, Z, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2016-08-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for dynamic regulation of the human 26S proteasome.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5TGA
 
 | |
5TGM
 
 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
5T1F
 
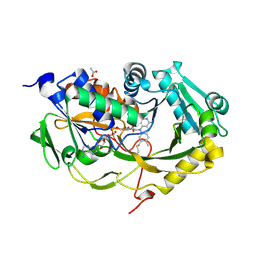 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5T1E
 
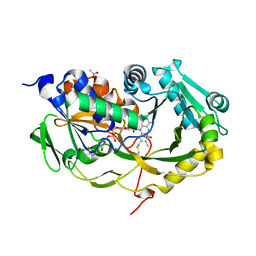 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
2RNL
 
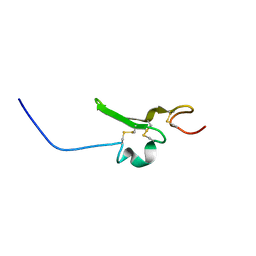 | | Solution structure of the EGF-like domain from human Amphiregulin | | Descriptor: | Amphiregulin | | Authors: | Qin, X, Hayashi, F, Terada, T, Shirouzu, M, Watanabe, S, Kigawa, T, Yabuta, N, Nojima, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-01-11 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the EGF-like domain from human Amphiregulin
To be Published
|
|
