6DSP
 
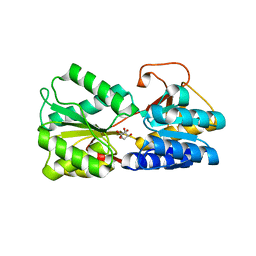 | | LsrB from Clostridium saccharobutylicum in complex with AI-2 | | Descriptor: | (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, Autoinducer 2-binding protein LsrB | | Authors: | Torcato, I.M, Xavier, K.B, Miller, S.T. | | Deposit date: | 2018-06-14 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Identification of novel autoinducer-2 receptors inClostridiareveals plasticity in the binding site of the LsrB receptor family.
J.Biol.Chem., 294, 2019
|
|
6DGO
 
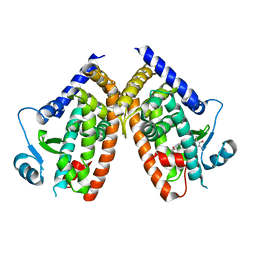 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Troglitazone | | Descriptor: | (5S)-5-[(4-{[(2R)-6-hydroxy-2,5,7,8-tetramethyl-3,4-dihydro-2H-1-benzopyran-2-yl]methoxy}phenyl)methyl]-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Quantitative structural assessment of graded receptor agonism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6DY0
 
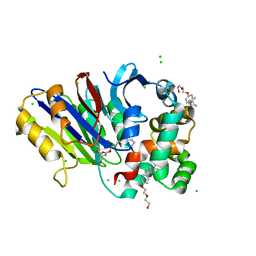 | | Rabbit N-acylethanolamine-hydrolyzing acid amidase (NAAA) covalently bound to beta-lactam inhibitor ARN726, in presence of Triton X-100 | | Descriptor: | (2S)-3-amino-2-{[(4-cyclohexylbutoxy)carbonyl]amino}propanethioic S-acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-{2-[4-(1,1,3,3-TETRAMETHYLBUTYL)PHENOXY]ETHOXY}ETHANOL, ... | | Authors: | Gorelik, A, Gebai, A, Illes, K, Piomelli, D, Nagar, B. | | Deposit date: | 2018-07-01 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.014 Å) | | Cite: | Molecular mechanism of activation of the immunoregulatory amidase NAAA.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DIR
 
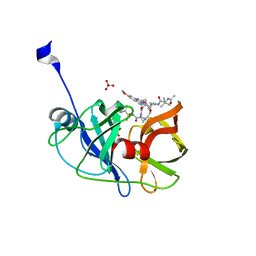 | | Crystal structure of HCV NS3/4A protease in complex with P4-P5-2 (AJ-67) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-3-methyl-L-valyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methyl cyclopropyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diaz acyclopentadecine-14a(5H)-carboxamide, NS3 protease, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6DIV
 
 | | Crystal structure of HCV NS3/4A D168A protease in complex with P4-P5-2 (AJ-67) | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(N-acetyl-3-methyl-L-valyl)amino]-2-[(7-methoxy-3-methylquinoxalin-2-yl)oxy]-N-[(1-methyl cyclopropyl)sulfonyl]-5,16-dioxo-1,2,3,6,7,8,9,10,11,13a,14,15,16,16a-tetradecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diaz acyclopentadecine-14a(5H)-carboxamide, GLYCEROL, NS3 protease, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-05-23 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design of Hepatitis C NS3/4A Protease Inhibitors Leveraging Untapped Regions of the Substrate Envelope
To Be Published
|
|
6DJ2
 
 | | HIV-1 protease with single mutation L76V in complex with Lopinavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Zhang, Y, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
5KBZ
 
 | | Structure of the PksA Product Template domain in complex with a phosphopantetheine mimetic | | Descriptor: | (14R)-14-hydroxy-15,15-dimethyl-1-[5-({[(5-methyl-1,2-oxazol-3-yl)methyl]sulfanyl}methyl)-1,2-oxazol-3-yl]-4,9,13-trioxo-2-thia-5,8,12-triazahexadecan-16-yl dihydrogen phosphate, Noranthrone synthase | | Authors: | Tsai, S.C, Burkart, M.D, Townsend, C.A, Barajas, J.F, Shakya, G, Topper, C.L, Moreno, G, Jackson, D.R, Rivera, H, La Clair, J.J, Vagstad, A. | | Deposit date: | 2016-06-03 | | Release date: | 2017-05-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Polyketide mimetics yield structural and mechanistic insights into product template domain function in nonreducing polyketide synthases.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KE2
 
 | | Crystal structure of SETDB1 Tudor domain in complex with inhibitor XST06472A | | Descriptor: | (3~{S})-~{N}-~{tert}-butyl-1,2,3,4-tetrahydroisoquinoline-3-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with inhibitor xst06472a
to be published
|
|
5KDF
 
 | | Inorganic pyrophosphatase from Mycobacterium tuberculosis in complex with inhibitor 6 and inorganic pyrophosphate | | Descriptor: | CALCIUM ION, Inorganic pyrophosphatase, PYROPHOSPHATE 2-, ... | | Authors: | Pang, A.H, Garzan, A, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Allosteric and Selective Inhibitors of Inorganic Pyrophosphatase from Mycobacterium tuberculosis.
ACS Chem. Biol., 11, 2016
|
|
6DL7
 
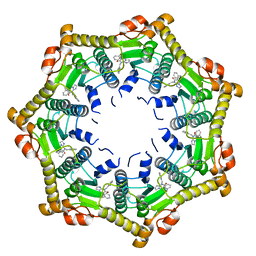 | | Human mitochondrial ClpP in complex with ONC201 (TIC10) | | Descriptor: | 7-benzyl-4-[(2-methylphenyl)methyl]-6,7,8,9-tetrahydroimidazo[1,2-a]pyrido[3,4-e]pyrimidin-5(4H)-one, ATP-dependent Clp protease proteolytic subunit, mitochondrial | | Authors: | Halgas, O, Zarabi, S.F, Schimmer, A, Pai, E.F. | | Deposit date: | 2018-05-31 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mitochondrial ClpP-Mediated Proteolysis Induces Selective Cancer Cell Lethality.
Cancer Cell, 35, 2019
|
|
5KGR
 
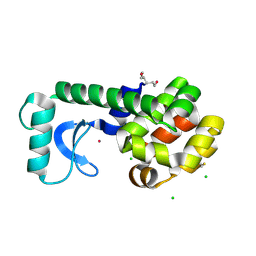 | | Spin-Labeled T4 Lysozyme Construct I9V1/V131V1 (30 days) | | Descriptor: | CHLORIDE ION, Endolysin, HEXANE-1,6-DIOL, ... | | Authors: | Balo, A.R, Feyrer, H, Ernst, O.P. | | Deposit date: | 2016-06-13 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.473 Å) | | Cite: | Toward Precise Interpretation of DEER-Based Distance Distributions: Insights from Structural Characterization of V1 Spin-Labeled Side Chains.
Biochemistry, 55, 2016
|
|
6DN3
 
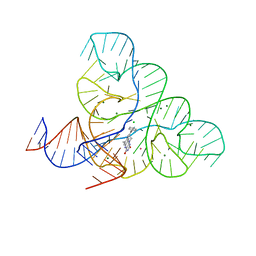 | | CRYSTAL STRUCTURE OF THE FMN RIBOSWITCH BOUND TO BRX1555 SPLIT RNA | | Descriptor: | 7,8-dimethyl-2,4-dioxo-10-(3-phenylpropyl)-1,2,3,4-tetrahydrobenzo[g]pteridin-10-ium, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Vicens, Q, Mondragon, E, Reyes, F.E, Berman, J, Kaur, H, Kells, K, Wickens, P, Wilson, J, Gadwood, R, Schostarez, H, Suto, R.K, Coish, P, Blount, K.F, Batey, R.T. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch.
ACS Chem. Biol., 13, 2018
|
|
4OH1
 
 | |
6U36
 
 | | PCSK9 in complex with a Fab and compound 14 | | Descriptor: | 2-fluoro-4-{[(1R)-6-(2-{4-[1-(4-methoxyphenyl)-5-methyl-6-oxo-1,6-dihydropyridazin-3-yl]-1H-1,2,3-triazol-1-yl}ethoxy)-1-methyl-1-{2-oxo-2-[(1,3-thiazol-2-yl)amino]ethyl}-1,2,3,4-tetrahydroisoquinolin-7-yl]oxy}benzoic acid, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Lu, J, Soisson, S. | | Deposit date: | 2019-08-21 | | Release date: | 2019-11-06 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9.
Cell Chem Biol, 27, 2020
|
|
1AIE
 
 | |
7M3K
 
 | |
6DTE
 
 | | GlcNAc-inspired cyclophellitol bound to NagZ | | Descriptor: | 2,2,2-trifluoro-N-[(1R,2R,3R,4R,5R,6R)-2,3,5,6-tetrahydroxy-4-(hydroxymethyl)cyclohexyl]acetamide, Beta-hexosaminidase | | Authors: | Mark, B.L, Winogrodzki, J.L. | | Deposit date: | 2018-06-15 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.929 Å) | | Cite: | A mechanism-based GlcNAc-inspired cyclophellitol inactivator of the peptidoglycan recycling enzyme NagZ reverses resistance to beta-lactams in Pseudomonas aeruginosa.
Chem. Commun. (Camb.), 54, 2018
|
|
5KPM
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD3731 | | Descriptor: | (4~{S})-3-(2,2-dimethylpropyl)-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KPL
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0705 | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
8JZB
 
 | | Langya Henipavirus attachment protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Yan, R.H, Guo, Y.Y. | | Deposit date: | 2023-07-05 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A special homotetrameric architecture of langya virus glycoprotein
To Be Published
|
|
5KPK
 
 | | Glycogen Synthase Kinase 3 beta Complexed with BRD0209 | | Descriptor: | (4~{S})-3-cyclopropyl-4,7,7-trimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Lakshminarasimhan, D, White, A, Nadupalli, A, Suto, R.K. | | Deposit date: | 2016-07-04 | | Release date: | 2018-03-14 | | Last modified: | 2018-03-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5L7H
 
 | | MCR IN COMPLEX WITH ligand | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 3-methyl-5,8-dioxa-17lambda-thia-4,18-diazatetracyclo[18.2.2.1,.0]pentacosa-1(22),2(6),3,9,11,13(25),20,23-octaene-17,17-dione, Mineralocorticoid receptor, ... | | Authors: | Xue, Y, Aagaard, A, Backstrom, S, Edman, K. | | Deposit date: | 2016-06-03 | | Release date: | 2016-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Based Drug Design of Mineralocorticoid Receptor Antagonists to Explore Oxosteroid Receptor Selectivity.
ChemMedChem, 12, 2017
|
|
5L7V
 
 | | Crystal Structure of BvGH123 with bond transition state analog Galthiazoline. | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
5LA7
 
 | | Crystal structure of human proheparanase, in complex with glucuronic acid configured aziridine probe JJB355 | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{S},6~{R})-2-(8-azidooctylamino)-3,4,5,6-tetrakis(oxidanyl)cyclohexane-1-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Jin, Y, Davies, G.J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Activity-based probes for functional interrogation of retaining beta-glucuronidases.
Nat. Chem. Biol., 13, 2017
|
|
