3S7R
 
 | |
4AMX
 
 | | CRYSTAL STRUCTURE OF THE GRACILARIOPSIS LEMANEIFORMIS ALPHA-1,4- GLUCAN LYASE Covalent Intermediate Complex with 5-fluoro-glucosyl- fluoride | | Descriptor: | 5-fluoro-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2012-03-14 | | Release date: | 2013-03-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
4J7G
 
 | | Crystal structure of EvaA, a 2,3-dehydratase in complex with dTDP-fucose and dTDP-rhamnose | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, EvaA 2,3-dehydratase, [[(2R,3S,5R)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3R,4S,5R,6R)-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Holden, H.M, Kubiak, R.L, Thoden, J.B. | | Deposit date: | 2013-02-13 | | Release date: | 2013-05-22 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of EvaA: A Paradigm for Sugar 2,3-Dehydratases.
Biochemistry, 52, 2013
|
|
5GM2
 
 | | Crystal structure of methyltransferase TleD complexed with SAH and teleocidin A1 | | Descriptor: | (2S,5S)-9-[(3R)-3,7-dimethylocta-1,6-dien-3-yl]-5-(hydroxymethyl)-1-methyl-2-(propan-2-yl)-1,2,4,5,6,8-hexahydro-3H-[1,4]diazonino[7,6,5-cd]indol-3-one, O-methylransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yu, F, Li, M.J, Xu, C.Y, Zhou, H, Sun, B, Wang, Z.J, Xu, Q, Xie, M.Y, Zuo, G, Huang, P, Wang, Q.S, He, J.H. | | Deposit date: | 2016-07-12 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure and enantioselectivity of terpene cyclization in SAM-dependent methyltransferase TleD
Biochem.J., 473, 2016
|
|
1FP4
 
 | | CRYSTAL STRUCTURE OF THE ALPHA-H195Q MUTANT OF NITROGENASE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE-MO-S CLUSTER, ... | | Authors: | Sorlie, M, Christiansen, J, Lemon, B.J, Peters, J.W, Dean, D.R, Hales, B.J. | | Deposit date: | 2000-08-30 | | Release date: | 2002-11-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic features and structure of the nitrogenase alpha-Gln195 MoFe protein
Biochemistry, 40, 2001
|
|
1T60
 
 | | Crystal structure of Type IV collagen NC1 domain from bovine lens capsule | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Vanacore, R.M, Shanmugasundararaj, S, Friedman, D.B, Bondar, O, Hudson, B.G, Sundaramoorthy, M. | | Deposit date: | 2004-05-05 | | Release date: | 2004-09-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links
J.Biol.Chem., 279, 2004
|
|
4ION
 
 | | Macrolepiota procera ricin B-like lectin (MPL) | | Descriptor: | GLYCEROL, Ricin B-like lectin | | Authors: | Renko, M, Zurga, S, Sabotic, J, Pohleven, J, Kos, J, Turk, D. | | Deposit date: | 2013-01-08 | | Release date: | 2013-11-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Macrolepiota procera ricin B-like lectin (MPL)
TO BE PUBLISHED
|
|
1MHJ
 
 | |
4ORO
 
 | |
5AG8
 
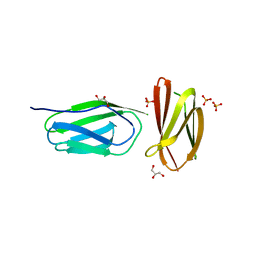 | | CRYSTAL STRUCTURE OF A MUTANT (665I6H) OF THE C-TERMINAL DOMAIN OF RGPB | | Descriptor: | GINGIPAIN R2, GLYCEROL, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Outer-Membrane Export Signal of Porphyromonas Gingivalis Type Ix Secretion System (T9Ss) is a Conserved C-Terminal Beta-Sandwich Domain.
Sci.Rep., 6, 2016
|
|
3L5N
 
 | |
2X2H
 
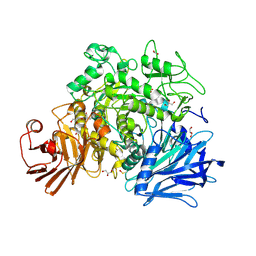 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase | | Descriptor: | ACETATE ION, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, CHLORIDE ION, ... | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
5AG9
 
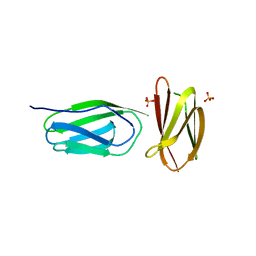 | | CRYSTAL STRUCTURE OF A MUTANT (665sXa) C-TERMINAL DOMAIN OF RGPB | | Descriptor: | Gingipain R2, SULFATE ION | | Authors: | de Diego, I, Ksiazek, M, Mizgalska, D, Golik, P, Szmigielski, B, Nowak, M, Nowakowska, Z, Potempa, B, Koneru, L, Nguyen, K.A, Enghild, J, Thogersen, I.B, Dubin, G, Gomis-Ruth, F.X, Potempa, J. | | Deposit date: | 2015-01-29 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
2WX5
 
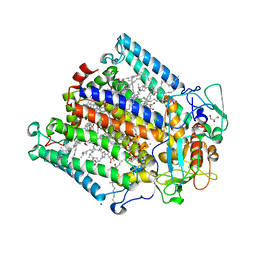 | | Hexa-coordination of a bacteriochlorophyll cofactor in the Rhodobacter sphaeroides reaction centre | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Marsh, M, Frolov, D, Crouch, L.I, Fyfe, P.K, Robert, B, van Grondelle, R, Jones, M.R, Hadfield, A.T. | | Deposit date: | 2009-11-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural and Spectroscopic Consequences of Hexa-Coordination of a Bacteriochlorophyll Cofactor in the Rhodobacter Sphaeroides Reaction Centre
Biochemistry, 49, 2010
|
|
7KDU
 
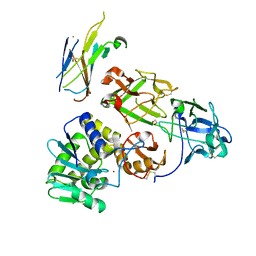 | | Ricin bound to VHH antibody V5E4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ricin chain A, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-09 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.809 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7KD0
 
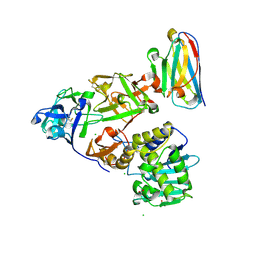 | | Ricin bound to VHH antibody V2C11 | | Descriptor: | 1,2-ETHANEDIOL, Anti-RON nanobody, CHLORIDE ION, ... | | Authors: | Rudolph, M.J. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.768 Å) | | Cite: | Structural Analysis of Toxin-Neutralizing, Single-Domain Antibodies that Bridge Ricin's A-B Subunit Interface.
J.Mol.Biol., 433, 2021
|
|
7YXW
 
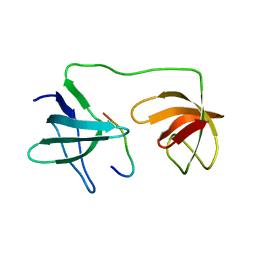 | | Structure of the p22phox A200G mutant in complex with p47phox peptide | | Descriptor: | Cytochrome b-245 light chain, Neutrophil cytosol factor 1 | | Authors: | Cukier, C.D, Vuillard, L.M, Komjati, B, Szlavik, Z. | | Deposit date: | 2022-02-16 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting NOX2 via p47/phox-p22/phox Inhibition with Novel Triproline Mimetics
Acs Med.Chem.Lett., 13, 2022
|
|
6GL9
 
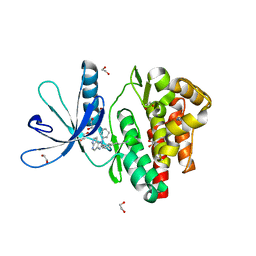 | | Crystal structure of JAK3 in complex with Compound 10 (FM475) | | Descriptor: | (~{E})-3-[3-(3-cyclohexyl-3,5,8,10-tetrazatricyclo[7.3.0.0^{2,6}]dodeca-1(9),2(6),4,7,11-pentaen-4-yl)phenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, 1-phenylurea, ... | | Authors: | Chaikuad, A, Forster, M, von Delft, F, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Laufer, S.A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development, Optimization, and Structure-Activity Relationships of Covalent-Reversible JAK3 Inhibitors Based on a Tricyclic Imidazo[5,4- d]pyrrolo[2,3- b]pyridine Scaffold.
J. Med. Chem., 61, 2018
|
|
1ABA
 
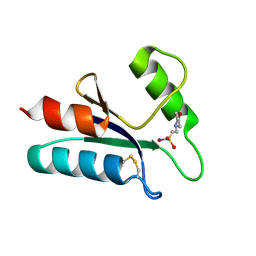 | | THE STRUCTURE OF OXIDIZED BACTERIOPHAGE T4 GLUTAREDOXIN (THIOREDOXIN). REFINEMENT OF NATIVE AND MUTANT PROTEINS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTAREDOXIN | | Authors: | Eklund, H, Ingelman, M, Soderberg, B.-O, Uhlin, T, Nordlund, P, Nikkola, M, Sonnerstam, U, Joelson, T, Petratos, K. | | Deposit date: | 1992-04-24 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of oxidized bacteriophage T4 glutaredoxin (thioredoxin). Refinement of native and mutant proteins.
J.Mol.Biol., 228, 1992
|
|
4RT7
 
 | | Crystal Structure of FLT3 with a small molecule inhibitor | | Descriptor: | 1-(5-tert-butyl-1,2-oxazol-3-yl)-3-(4-{7-[2-(morpholin-4-yl)ethoxy]imidazo[2,1-b][1,3]benzothiazol-2-yl}phenyl)urea, Receptor-type tyrosine-protein kinase FLT3 | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2014-11-13 | | Release date: | 2015-04-22 | | Last modified: | 2015-06-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Characterizing and Overriding the Structural Mechanism of the Quizartinib-Resistant FLT3 "Gatekeeper" F691L Mutation with PLX3397.
Cancer Discov, 5, 2015
|
|
5HFS
 
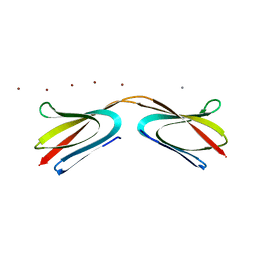 | | CRYSTAL STRUCTURE OF C-TERMINAL DOMAIN OF CARGO PROTEINS OF TYPE IX SECRETION SYSTEM | | Descriptor: | CALCIUM ION, Gingipain R2, ZINC ION | | Authors: | Golik, P, Szmigielski, B, Ksiazek, M, Nowakowska, Z, Mizgalska, D, Nowak, M, Dubin, G, Potempa, J. | | Deposit date: | 2016-01-07 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal beta-sandwich domain.
Sci Rep, 6, 2016
|
|
1UV6
 
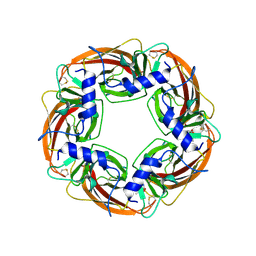 | | X-ray structure of acetylcholine binding protein (AChBP) in complex with carbamylcholine | | Descriptor: | 2-[(AMINOCARBONYL)OXY]-N,N,N-TRIMETHYLETHANAMINIUM, ACETYLCHOLINE-BINDING PROTEIN | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-01-15 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
2NV4
 
 | | Crystal structure of UPF0066 protein AF0241 in complex with S-adenosylmethionine. Northeast Structural Genomics Consortium target GR27 | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, UPF0066 protein AF_0241 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-11-10 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional insights from structural genomics.
J.Struct.Funct.Genom., 8, 2007
|
|
1N3N
 
 | | Crystal structure of a mycobacterial hsp60 epitope with the murine class I MHC molecule H-2Db | | Descriptor: | BETA-2-MICROGLOBULIN, H-2 CLASS I HISTOCOMPATIBILITY ANTIGEN, D-B ALPHA CHAIN, ... | | Authors: | Ciatto, C, Capitani, G, Tissot, A.C, Pecorari, F, Pluckthun, A, Grutter, M.G. | | Deposit date: | 2002-10-29 | | Release date: | 2003-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural analysis of mycobacterial
and murine hsp60 epitopes in
complex with the class I MHC
molecule H-2D(b)
FEBS Lett., 543, 2003
|
|
3POT
 
 | |
