4U76
 
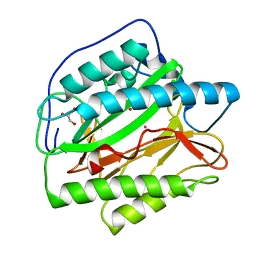 | | HsMetAP (F309M) holo form | | Descriptor: | COBALT (II) ION, GLYCEROL, Methionine aminopeptidase 1, ... | | Authors: | Arya, T, Addlagatta, A. | | Deposit date: | 2014-07-30 | | Release date: | 2015-03-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Identification of the Molecular Basis of Inhibitor Selectivity between the Human and Streptococcal Type I Methionine Aminopeptidases
J.Med.Chem., 58, 2015
|
|
4UAI
 
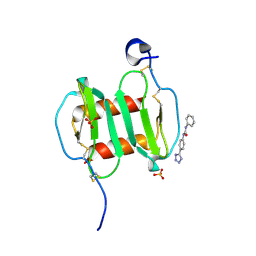 | | Crystal structure of CXCL12 in complex with inhibitor | | Descriptor: | 1-phenyl-3-[4-(1H-tetrazol-5-yl)phenyl]urea, SULFATE ION, Stromal cell-derived factor 1 | | Authors: | Smith, E.W, Chen, Y. | | Deposit date: | 2014-08-09 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Novel Small Molecule Ligand Bound to the CXCL12 Chemokine.
J.Med.Chem., 57, 2014
|
|
4U73
 
 | |
5OMG
 
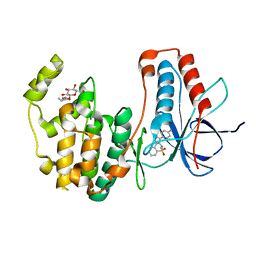 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXP4M12 | | Descriptor: | 3-(4-fluorophenyl)-4-methyl-1~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
4U7K
 
 | | Crystal structure of the Clostridium histolyticum colH collagenase polycystic kidney disease-like domain 2a in the presence of calcium at 1.9 Angstrom resolution | | Descriptor: | CALCIUM ION, ColH protein | | Authors: | Bauer, R, Janowska, K, Sakon, J, Matsushita, O, Latimer, E. | | Deposit date: | 2014-07-31 | | Release date: | 2015-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of three polycystic kidney disease-like domains from Clostridium histolyticum collagenases ColG and ColH.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UBQ
 
 | | Crystal Structure of IMP-2 Metallo-beta-Lactamase from Acinetobacter spp. | | Descriptor: | ACETATE ION, Beta-lactamase, ZINC ION | | Authors: | Yamaguchi, Y, Matsueda, S, Matsunaga, K, Takashio, N, Toma-Fukai, S, Yamagata, Y, Shibata, N, Wachino, J, Shibayama, K, Arakawa, Y, Kurosaki, H. | | Deposit date: | 2014-08-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of IMP-2 metallo-beta-lactamase from Acinetobacter spp.: comparison of active-site loop structures between IMP-1 and IMP-2.
Biol.Pharm.Bull., 38, 2015
|
|
5OMH
 
 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXH11 | | Descriptor: | 1-(3-chlorophenyl)-3-methyl-4~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
7VR6
 
 | | Crystal structure of MlaC from Escherichia coli in quasi-open state | | Descriptor: | 1,2-ETHANEDIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Intermembrane phospholipid transport system binding protein MlaC | | Authors: | Dutta, A, Kanaujia, S.P. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MlaC belongs to a unique class of non-canonical substrate-binding proteins and follows a novel phospholipid-binding mechanism.
J.Struct.Biol., 214, 2022
|
|
4UR9
 
 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4V5X
 
 | | The cryo-EM structure of a 3D DNA-origami object | | Descriptor: | SCAFFOLD STRAND,SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Bai, X.C, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2012-10-09 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Cryo-Em Structure of a 3D DNA-Origami Object.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8YTE
 
 | | Crystal Structure of TrkA D5 domain in complex with macrocyclic peptide | | Descriptor: | 1,2-ETHANEDIOL, AMINOMETHYLAMIDE, High affinity nerve growth factor receptor, ... | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
8Z2C
 
 | |
5N67
 
 | |
9JGC
 
 | | Crystal structure of Nep1 in complex with adenosine from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, CHLORIDE ION, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
9JGB
 
 | | Crystal structure of Nep1 from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ribosomal RNA small subunit methyltransferase Nep1, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
9JGD
 
 | | Crystal structure of Nep1 in complex with 5'-methylthioadenosine from Pyrococcus horikoshii OT3 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, ... | | Authors: | Saha, S, Kanaujia, S.P. | | Deposit date: | 2024-09-06 | | Release date: | 2025-03-12 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the ribosome assembly factor Nep1, an N1-specific pseudouridine methyltransferase, reveals mechanistic insights.
Febs J., 292, 2025
|
|
8Z0G
 
 | | Crystal structure of NeIle complexed with isoleucine | | Descriptor: | CHLORIDE ION, GLYCEROL, ISOLEUCINE, ... | | Authors: | Samygina, V.R, Subach, O.M, Vlaskina, A.V, Gabdukhakov, A, Subach, F.V. | | Deposit date: | 2024-04-09 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | NeIle, a Genetically Encoded Indicator for Branched-Chain Amino Acids Based on mNeonGreen Fluorescent Protein and LIVBP Protein.
ACS Sens, 9, 2024
|
|
8Z2X
 
 | | Crystal structure of exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) as a complex with cellobiose | | Descriptor: | Glucan 1,3-beta-glucosidase A, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 599, 2025
|
|
7JN9
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide QEHTGSQLRIAAYGP | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8Z2W
 
 | | Crystal structure of apo- exo-beta-(1,3)-glucanase from Aspergillus oryzae (AoBgl) | | Descriptor: | 2-methylpropylphosphonic acid, Glucan 1,3-beta-glucosidase A, SODIUM ION | | Authors: | Banerjee, B, Kamale, C.K, Suryawanshi, A.B, Bhaumik, P. | | Deposit date: | 2024-04-13 | | Release date: | 2024-11-06 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Aspergillus oryzae exo-beta-(1,3)-glucanase reveal insights into oligosaccharide binding, recognition, and hydrolysis.
Febs Lett., 599, 2025
|
|
8YTD
 
 | | Crystal Structure of TrkA D5 domain in complex with two different macrocyclic peptides | | Descriptor: | 1,2-ETHANEDIOL, High affinity nerve growth factor receptor, Macrocyclic Peptide | | Authors: | Yamada, T, Mihara, K, Ueda, T, Yamauchi, D, Shimizu, M, Ando, A, Mayumi, K, Nakata, Z, Mikamiyama, H. | | Deposit date: | 2024-03-25 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Hit to Lead Optimization of Macrocyclic Peptides as Novel Tropomyosin Receptor Kinase A Antagonists.
J.Med.Chem., 67, 2024
|
|
7JMM
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RAKNIILLSR | | Descriptor: | Alkaline phosphatase, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-02 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JN8
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGNTLVIVSR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JNE
 
 | | Crystal structure of the substrate-binding domain of E. coli DnaK in complex with the peptide RGSQLRIASR | | Descriptor: | Alkaline phosphatase peptide, Chaperone protein DnaK, SULFATE ION | | Authors: | Jansen, R.M, Ozden, C, Gierasch, L.M, Garman, S.C. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Selective promiscuity in the binding of E. coli Hsp70 to an unfolded protein.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5OAK
 
 | |
