9G60
 
 | |
1EOV
 
 | | FREE ASPARTYL-TRNA SYNTHETASE (ASPRS) (E.C. 6.1.1.12) FROM YEAST | | Descriptor: | ASPARTYL-TRNA SYNTHETASE | | Authors: | Sauter, C, Lorber, B, Cavarelli, J, Moras, D, Giege, R. | | Deposit date: | 2000-03-24 | | Release date: | 2000-09-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free yeast aspartyl-tRNA synthetase differs from the tRNA(Asp)-complexed enzyme by structural changes in the catalytic site, hinge region, and anticodon-binding domain.
J.Mol.Biol., 299, 2000
|
|
5LM4
 
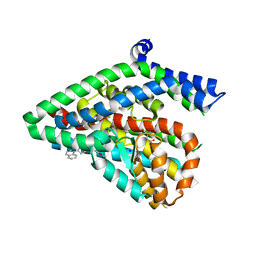 | | Structure of the thermostalilized EAAT1 cryst-II mutant in complex with L-ASP and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J, Assal, R, Legrand, P, Reyes, N. | | Deposit date: | 2016-07-29 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and allosteric inhibition of excitatory amino acid transporter 1.
Nature, 544, 2017
|
|
8Q1X
 
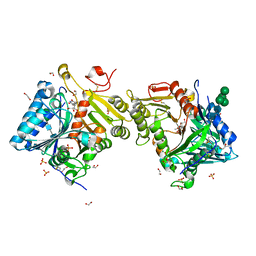 | | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5'-3' exonuclease PLD3, ... | | Authors: | Roske, Y, Daumke, O, Damme, M. | | Deposit date: | 2023-08-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of PLD3 reveals insights into the mechanism of lysosomal 5' exonuclease-mediated nucleic acid degradation.
Nucleic Acids Res., 52, 2024
|
|
9G5F
 
 | |
1EPG
 
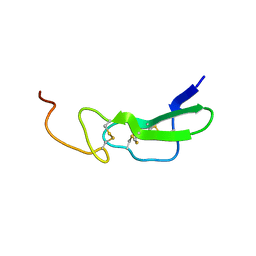 | |
5LMQ
 
 | | Structure of bacterial 30S-IF1-IF3-mRNA-tRNA translation pre-initiation complex, open form (state-2A) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Hussain, T, Llacer, J.L, Wimberly, B.T, Ramakrishnan, V. | | Deposit date: | 2016-08-01 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Large-Scale Movements of IF3 and tRNA during Bacterial Translation Initiation.
Cell, 167, 2016
|
|
1EPP
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-130,693 (MAS PHE LYS+MTF STA MBA) | | Descriptor: | ENDOTHIAPEPSIN, N-(dimethylsulfamoyl)-L-phenylalanyl-N-[(1S,2S)-2-hydroxy-4-{[(2S)-2-methylbutyl]amino}-1-(2-methylpropyl)-4-oxobutyl]-N~6~-(methylcarbamothioyl)-L-lysinamide, SULFATE ION | | Authors: | Wallace, B.A, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analyses of ligand binding in five endothiapepsin crystal complexes and their use in the design and evaluation of novel renin inhibitors.
J.Med.Chem., 36, 1993
|
|
1EQ5
 
 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
5LNX
 
 | |
9G5R
 
 | |
1EQE
 
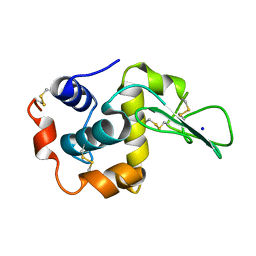 | | CRYSTAL STRUCTURES OF SALT BRIDGE MUTANTS OF HUMAN LYSOZYME | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Tsuchimori, K, Yamagata, Y, Yutani, K. | | Deposit date: | 2000-04-04 | | Release date: | 2000-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of salt bridges near the surface of a protein to the conformational stability.
Biochemistry, 39, 2000
|
|
5LOX
 
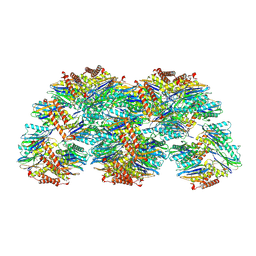 | |
5M32
 
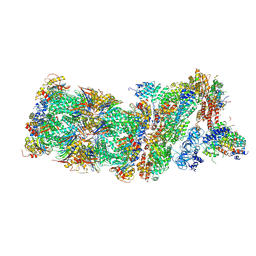 | | Human 26S proteasome in complex with Oprozomib | | Descriptor: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | Authors: | Haselbach, D, Schrader, J, Lambrecht, F, Henneberg, F, Chari, A, Stark, H. | | Deposit date: | 2016-10-14 | | Release date: | 2017-07-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs.
Nat Commun, 8, 2017
|
|
1EQK
 
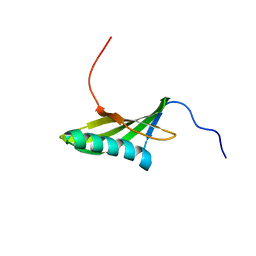 | | SOLUTION STRUCTURE OF ORYZACYSTATIN-I, A CYSTEINE PROTEINASE INHIBITOR OF THE RICE, ORYZA SATIVA L. JAPONICA | | Descriptor: | ORYZACYSTATIN-I | | Authors: | Nagata, K, Kudo, N, Abe, K, Arai, S, Tanokura, M. | | Deposit date: | 2000-04-05 | | Release date: | 2001-01-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica.
Biochemistry, 39, 2000
|
|
1ERW
 
 | | HUMAN THIOREDOXIN DOUBLE MUTANT WITH CYS 32 REPLACED BY SER AND CYS 35 REPLACED BY SER | | Descriptor: | THIOREDOXIN | | Authors: | Weichsel, A, Gasdaska, J.R, Powis, G, Montfort, W.R. | | Deposit date: | 1996-02-07 | | Release date: | 1996-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer.
Structure, 4, 1996
|
|
5LS7
 
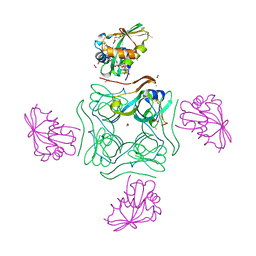 | | Complex of wild type E. coli alpha aspartate decarboxylase with its processing factor PanZ | | Descriptor: | ACETYL COENZYME *A, Aspartate 1-decarboxylase, CARBON DIOXIDE, ... | | Authors: | Monteiro, D.C.F, Webb, M.E, Pearson, A.R. | | Deposit date: | 2016-08-22 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | The Mechanism of Regulation of Pantothenate Biosynthesis by the PanD-PanZAcCoA Complex Reveals an Additional Mode of Action for the Antimetabolite N-Pentyl Pantothenamide (N5-Pan).
Biochemistry, 56, 2017
|
|
1ESP
 
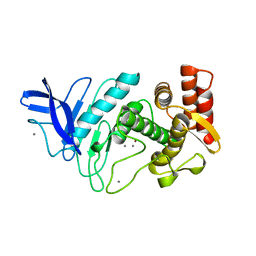 | | NEUTRAL PROTEASE MUTANT E144S | | Descriptor: | CALCIUM ION, NEUTRAL PROTEASE MUTANT E144S, ZINC ION | | Authors: | Litster, S.A, Wetmore, D.R, Roche, R.S, Codding, P.W. | | Deposit date: | 1995-08-11 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | E144S active-site mutant of the Bacillus cereus thermolysin-like neutral protease at 2.8 A resolution.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1ETE
 
 | |
9G6L
 
 | | Xylose Isomerase collected at 20C using time-resolved serial synchrotron crystallography with Glucose at 60 seconds | | Descriptor: | D-glucose, MAGNESIUM ION, Xylose isomerase, ... | | Authors: | Schulz, E.C, Prester, A, Stetten, D.V, Gore, G, Hatton, C.E, Bartels, K, Leimkohl, J.P, Schikora, H, Ginn, H.M, Tellkamp, F, Mehrabi, P. | | Deposit date: | 2024-07-18 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the modulation of enzyme kinetics by multi-temperature, time-resolved serial crystallography.
Nat Commun, 16, 2025
|
|
1EU2
 
 | |
5LSY
 
 | | Structure of the Epigenetic Oncogene MMSET and inhibition by N-Alkyl Sinefungin Derivatives | | Descriptor: | Histone-lysine N-methyltransferase SETD2, THIOCYANATE ION, ZINC ION, ... | | Authors: | Tisi, D, Pathuri, P, Heightman, T. | | Deposit date: | 2016-09-05 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of the Epigenetic Oncogene MMSET and Inhibition by N-Alkyl Sinefungin Derivatives.
ACS Chem. Biol., 11, 2016
|
|
1EVH
 
 | |
5M4Y
 
 | |
5M5B
 
 | | Crystal structure of Zika virus NS5 methyltransferase | | Descriptor: | CHLORIDE ION, GLYCEROL, NS5 methyltransferase, ... | | Authors: | Barral, K, Ortiz Lombardia, M, Coutard, B, Decroly, E, Lichiere, J. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Zika Virus Methyltransferase: Structure and Functions for Drug Design Perspectives.
J. Virol., 91, 2017
|
|
