6CL1
 
 | |
2BHM
 
 | | Crystal structure of VirB8 from Brucella suis | | Descriptor: | TYPE IV SECRETION SYSTEM PROTEIN VIRB8 | | Authors: | Bayliss, R, Baron, C, Waksman, G. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BJ6
 
 | | Crystal Structure of a decameric HNA-RNA hybrid | | Descriptor: | 5'-R(*GP*GP*CP*AP*UP*UP*AP*CP*GP*GP)-3', SULFATE ION, SYNTHETIC HNA | | Authors: | Maier, T, Przylas, I, Straeter, N, Herdewijn, P, Saenger, W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid
J.Am.Chem.Soc., 127, 2005
|
|
2BON
 
 | | Structure of an Escherichia coli lipid kinase (YegS) | | Descriptor: | LIPID KINASE, MAGNESIUM ION | | Authors: | Bakali, H.M, Johnson, K.A, Hallberg, B.M, Herman, M.D, Nordlund, P. | | Deposit date: | 2005-04-12 | | Release date: | 2006-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Yegs, a Homologue to the Mammalian Diacylglycerol Kinases, Reveals a Novel Regulatory Metal Binding Site.
J.Biol.Chem., 282, 2007
|
|
2BRU
 
 | | Complex of the domain I and domain III of Escherichia coli transhydrogenase | | Descriptor: | NAD(P) TRANSHYDROGENASE SUBUNIT ALPHA, NAD(P) TRANSHYDROGENASE SUBUNIT BETA, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Johansson, T, Pedersen, A, Leckner, J, Karlsson, B.G. | | Deposit date: | 2005-05-11 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of a Transient Complex by NMR Using Paramagnetic Distance Restraints - the Complex of the Soluble Domains of Escherichia Coli Transhydrogenase
To be Published
|
|
6D0H
 
 | |
2BUS
 
 | |
2BVE
 
 | | Structure of the N-terminal of Sialoadhesin in complex with 2-Phenyl- Prop5Ac | | Descriptor: | SIALOADHESIN, benzyl 3,5-dideoxy-5-(propanoylamino)-D-glycero-alpha-D-galacto-non-2-ulopyranosidonic acid | | Authors: | Zaccai, N.R, May, A.P, Robinson, R.C, Burtnick, L.D, Crocker, P, Brossmer, R, Kelm, S, Jones, E.Y. | | Deposit date: | 2005-06-27 | | Release date: | 2006-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and in Silico Analysis of the Sialoside-Binding Characteristics of the Siglec Sialoadhesin.
J.Mol.Biol., 365, 2007
|
|
2BVO
 
 | | Structures of Three HIV-1 HLA-B5703-Peptide Complexes and Identification of Related HLAs Potentially Associated with Long-Term Non-Progression | | Descriptor: | BETA-2-MICROGLOBULIN, GAG PROTEIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Stewart-Jones, G.B, Gillespie, G, Overton, I.M, Kaul, R, Roche, P, Mcmichael, A.J, Rowland-Jones, S, Jones, E.Y. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structures of three HIV-1 HLA-B*5703-peptide complexes and identification of related HLAs potentially associated with long-term nonprogression.
J Immunol., 175, 2005
|
|
2BV9
 
 | | HOW FAMILY 26 GLYCOSIDE HYDROLASES ORCHESTRATE CATALYSIS ON DIFFERENT POLYSACCHARIDES. STRUCTURE AND ACTIVITY OF A CLOSTRIDIUM THERMOCELLUM LICHENASE, CtLIC26A | | Descriptor: | ENDOGLUCANASE H | | Authors: | Taylor, E.J, Goyal, A, Guerreiro, C.I.P.D, Prates, J.A.M, Money, V.A, Ferry, N, Morland, C, Planas, A, Macdonald, J.A, Stick, R.V, Gilbert, H.J, Fontes, C.M.G.A, Davies, G.J. | | Deposit date: | 2005-06-23 | | Release date: | 2005-06-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How Family 26 Glycoside Hydrolases Orchestrate Catalysis on Different Polysaccharides: Structure and Activity of a Clostridium Thermocellum Lichenase, Ctlic26A.
J.Biol.Chem., 280, 2005
|
|
6D16
 
 | | Crystal structure of KPC-2 complexed with compound 2 | | Descriptor: | Carbapenem-hydrolyzing beta-lactamase KPC, GLYCEROL, [(7-methoxy-2-oxo-2H-1-benzopyran-4-yl)methyl]phosphonic acid | | Authors: | Pemberton, O.A, Chen, Y. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Heteroaryl Phosphonates as Noncovalent Inhibitors of Both Serine- and Metallocarbapenemases.
J.Med.Chem., 62, 2019
|
|
2C53
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | Authors: | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2BQI
 
 | |
6CTN
 
 | |
6D1Y
 
 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 2,4-dichloro-N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)benzamide Inhibitor | | Descriptor: | 2,4-dichloro-N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)benzamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
2BEW
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXYPHENYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BGW
 
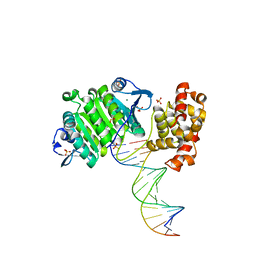 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BK1
 
 | | The pore structure of pneumolysin, obtained by fitting the alpha carbon trace of perfringolysin O into a cryo-EM map | | Descriptor: | PERFRINGOLYSIN O | | Authors: | Tilley, S.J, Orlova, E.V, Gilbert, R.J.C, Andrew, P.W, Saibil, H.R. | | Deposit date: | 2005-02-10 | | Release date: | 2005-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (29 Å) | | Cite: | Structural Basis of Pore Formation by the Bacterial Toxin Pneumolysin
Cell(Cambridge,Mass.), 121, 2005
|
|
2BW9
 
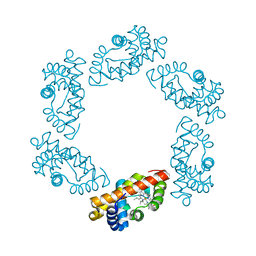 | | Laue Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Ligand Migration Pathway and Protein Dynamics in Myoglobin: A Time-Resolved Crystallographic Study on L29W Mbco.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BKD
 
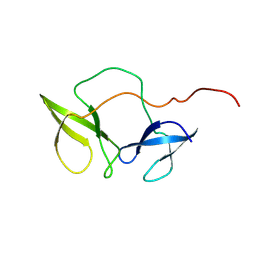 | | Structure of the N-terminal domain of Fragile X Mental Retardation Protein | | Descriptor: | Fragile X messenger ribonucleoprotein 1 | | Authors: | Ramos, A, Hollingworth, D, Adinolfi, S, Castets, M, Kelly, G, Frenkiel, T.A, Bardoni, B, Pastore, A. | | Deposit date: | 2005-02-15 | | Release date: | 2006-01-18 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction.
Structure, 14, 2006
|
|
2BSP
 
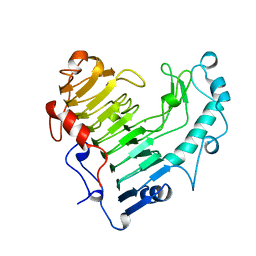 | | BACILLUS SUBTILIS PECTATE LYASE R279K MUTANT | | Descriptor: | CALCIUM ION, PROTEIN (PECTATE LYASE) | | Authors: | Pickersgill, R. | | Deposit date: | 1998-07-31 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Conserved Arginine Proximal to the Essential Calcium of Bacillus Subtilis Pectate Lyase Stabilizes the Transition State
To be Published
|
|
6D35
 
 | | Crystal structure of Xenopus Smoothened in complex with cholesterol | | Descriptor: | CHOLESTEROL, Smoothened,Soluble cytochrome b562,Smoothened | | Authors: | Huang, P, Zheng, S, Kim, Y, Kruse, A.C, Salic, A. | | Deposit date: | 2018-04-14 | | Release date: | 2018-05-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis of Smoothened Activation in Hedgehog Signaling.
Cell, 174, 2018
|
|
2BXV
 
 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 2-({[3-FLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENT-1-ENE-1-CARBOXYLIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Baumgartner, R, Walloschek, M, Karlik, M, Gotschlich, A, Tasler, S, Mies, J, Leban, J. | | Deposit date: | 2005-07-27 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J. Med. Chem., 49, 2006
|
|
6CVS
 
 | | Human Aprataxin (Aptx) L248M bound to DNA, AMP and Zn product | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, BETA-MERCAPTOETHANOL, ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
6D3P
 
 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | Authors: | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
