5GRJ
 
 | | Crystal structure of human PD-L1 with monoclonal antibody avelumab | | Descriptor: | Programmed cell death 1 ligand 1, avelumab H chain, avelumab L chain | | Authors: | Liu, K, Tan, S, Chai, Y, Chen, D, Song, H, Zhang, C.W.-H, Shi, Y, Liu, J, Tan, W, Lyu, J, Gao, S, Yan, J, Qi, J, Gao, G.F. | | Deposit date: | 2016-08-11 | | Release date: | 2016-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.206 Å) | | Cite: | Structural basis of anti-PD-L1 monoclonal antibody avelumab for tumor therapy.
Cell Res., 27, 2017
|
|
5IMK
 
 | | Nanobody targeting human Vsig4 in Spacegroup C2 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IML
 
 | | Nanobody targeting human Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, V-set and immunoglobulin domain-containing protein 4 | | Authors: | Wen, Y. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IUS
 
 | | Crystal structure of human PD-L1 in complex with high affinity PD-1 mutant | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Pascolutti, R, Sun, X, Kao, J, Maute, R, Ring, A.M, Bowman, G.R, Kruse, A.C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.889 Å) | | Cite: | Structure and Dynamics of PD-L1 and an Ultra-High-Affinity PD-1 Receptor Mutant.
Structure, 24, 2016
|
|
5JDR
 
 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
4F8T
 
 | | Crystal Structure of the Human BTN3A3 Ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A3 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell response by CD277/Butryophilin (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F9L
 
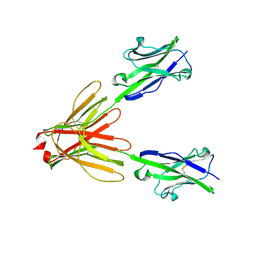 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 20.1 Single Chain Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 20.1 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1404 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4FMF
 
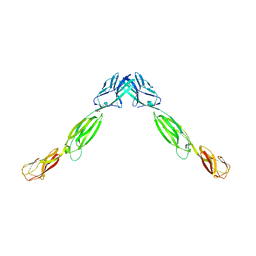 | |
4FMK
 
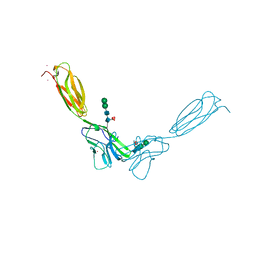 | | Crystal structure of mouse nectin-2 extracellular fragment D1-D2 | | Descriptor: | CADMIUM ION, Poliovirus receptor-related protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-17 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FQP
 
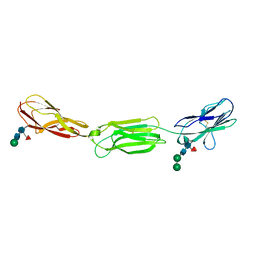 | | Crystal structure of human Nectin-like 5 full ectodomain (D1-D3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, ... | | Authors: | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-25 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4F8Q
 
 | | Crystal Structure of the Human BTN3A2 Ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A2 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3789 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4FN0
 
 | | Crystal structure of mouse nectin-2 extracellular fragment D1-D2, 2nd crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor-related protein 2, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-18 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FOM
 
 | | Crystal structure of human nectin-3 full ectodomain (D1-D3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor-related protein 3, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Jin, X, Brasch, J, Shapiro, L. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.93 Å) | | Cite: | Nectin ectodomain structures reveal a canonical adhesive interface.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4FS0
 
 | |
4FRW
 
 | |
4F9P
 
 | | Crystal Structure of the Human BTN3A1 Ectodomain in Complex with the 103.2 Single Chain Antibody | | Descriptor: | 103.2 anti-BTN3A1 antibody fragment, Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.519 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4F80
 
 | | Crystal Structure of the human BTN3A1 ectodomain | | Descriptor: | Butyrophilin subfamily 3 member A1 | | Authors: | Palakodeti, A, Sandstrom, A, Sundaresan, L, Harly, C, Nedellec, S, Olive, D, Scotet, E, Bonneville, M, Adams, E.J. | | Deposit date: | 2012-05-16 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | The molecular basis for modulation of human V(gamma)9V(delta)2 T cell responses by CD277/Butyrophilin-3 (BTN3A)-specific antibodies
J.Biol.Chem., 287, 2012
|
|
4HH8
 
 | | Crystal structure of bovine butyrophilin | | Descriptor: | Butyrophilin subfamily 1 member A1 | | Authors: | Eichinger, A, Skerra, A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-11-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The extracellular region of bovine butyrophilin exhibits high structural similarity to human myelin oligodendrocyte glycoprotein
To be Published
|
|
2JJT
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE SUBSTRATE 1 | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
2JJS
 
 | | Structure of human CD47 in complex with human signal regulatory protein (SIRP) alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IODIDE ION, LEUKOCYTE SURFACE ANTIGEN CD47, ... | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47.
Mol. Cell, 31, 2008
|
|
4Z18
 
 | | CRYSTAL STRUCTURE OF HUMAN PD-L1 | | Descriptor: | CHLORIDE ION, Programmed cell death 1 ligand 1 | | Authors: | Fedorov, A.A, Fedorov, E.V, Samantha, D, Hillerich, B, Seidel, R.D, Almo, S.C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | CRYSTAL STRUCTURE OF HUMAN PD-L1
To Be Published
|
|
5B22
 
 | | Dimer structure of murine Nectin-3 D1D2 | | Descriptor: | Nectin-3, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Takebe, K, Sangawa, T, Katsutani, T, Narita, H, Suzuki, M. | | Deposit date: | 2015-12-28 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Dimer structure of murine Nectin-3 D1D2
To Be Published
|
|
7WI0
 
 | | SARS-CoV-2 Omicron variant spike in complex with three human neutralizing antibodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c1437_light_IGLV1-40_IGLJ1, IG c934_light_IGKV1-5_IGKJ1, ... | | Authors: | Zheng, Q, Li, S, Sun, H, Zheng, Z, Wang, S. | | Deposit date: | 2022-01-01 | | Release date: | 2022-06-22 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Three SARS-CoV-2 antibodies provide broad and synergistic neutralization against variants of concern, including Omicron.
Cell Rep, 39, 2022
|
|
7WP5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spikeprotein S1, VacW-209 heavy chain, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
7WP0
 
 | | Cryo-EM structure of SARS-CoV-2 Delta S6P trimer in complex with neutralizing antibody VacW-209 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IG c335_light_IGLV1-40_IGLJ3, Spike protein S1, ... | | Authors: | Zheng, Q, Sun, H, Ju, B, Zhang, Z, Li, S, Xia, N. | | Deposit date: | 2022-01-22 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Immune escape by SARS-CoV-2 Omicron variant and structural basis of its effective neutralization by a broad neutralizing human antibody VacW-209.
Cell Res., 32, 2022
|
|
