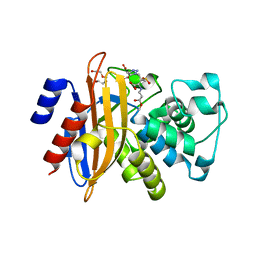
3LEZ
 
 | |
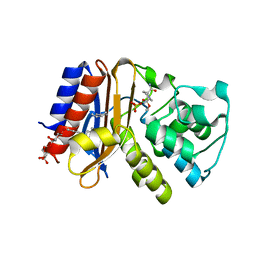
4ZAM
 
 | |
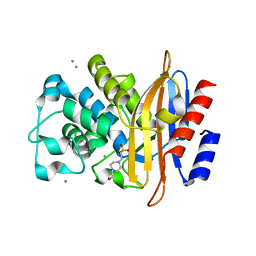
4A5R
 
 | | Crystal structure of class A beta-lactamase from Bacillus licheniformis BS3 with tazobactam | | Descriptor: | BETA-LACTAMASE, CARBON DIOXIDE, CITRIC ACID, ... | | Authors: | Power, P, Sauvage, E, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2011-10-28 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Class a Beta-Lactamase from Bacillus Licheniformis Inhibited by Tazobactam
To be Published
|
|
4BD1
 
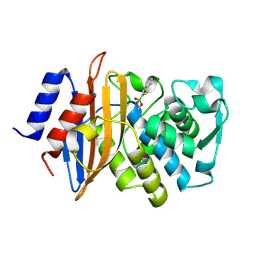 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Weiss, K.L, Standaert, R.F, Ostermann, A, Schrader, T.E, Ng, J.D, Coates, L. | | Deposit date: | 2012-10-04 | | Release date: | 2013-01-16 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.002 Å) | | Cite: | Neutron and X-Ray Crystal Structures of a Perdeuterated Enzyme Inhibitor Complex Reveal the Catalytic Proton Network of the Toho-1 Beta-Lactamase for the Acylation Reaction.
J.Biol.Chem., 288, 2013
|
|
4ZJ1
 
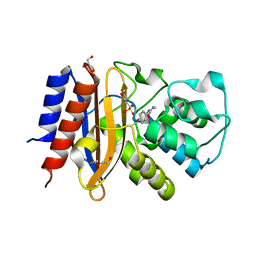 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli : V216AcrF mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Beta-lactamase TEM, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3JYI
 
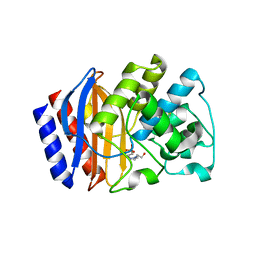 | | Structural and biochemical evidence that a TEM-1 {beta}-lactamase Asn170Gly active site mutant acts via substrate-assisted catalysis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase TEM, PHOSPHATE ION | | Authors: | Brown, N.G, Palzkill, T.G, Prasad, B.V.V, Shanker, S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structural and biochemical evidence that a TEM-1 beta-lactamase N170G active site mutant acts via substrate-assisted catalysis
J.Biol.Chem., 284, 2009
|
|
4ZBE
 
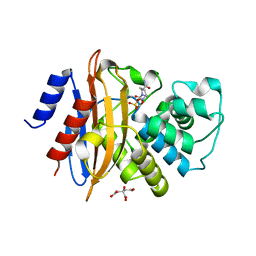 | |
4ZJ2
 
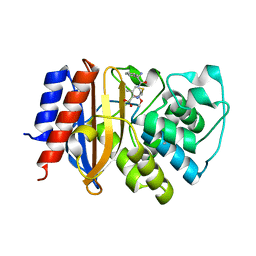 | | Crystal Structure of p-acrylamido-phenylalanine modified TEM1 beta-lactamase from Escherichia coli :E166N mutant | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, C.S, Schultz, P.G. | | Deposit date: | 2015-04-28 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4C3Q
 
 | | Neutron structure of a perdeuterated Toho-1 R274N R276N double mutant Beta-lactamase in complex with a fully deuterated boronic acid (BZB) at 100K | | Descriptor: | BENZO[B]THIOPHENE-2-BORONIC ACID, BETA-LACTAMASE TOHO-1 | | Authors: | Coates, L, Tomanicek, S.J, Schrader, T, Weiss, K.L, Ng, J.D, Ostermann, A. | | Deposit date: | 2013-08-26 | | Release date: | 2014-07-09 | | Last modified: | 2017-03-22 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Cryogenic Neutron Protein Crystallography: Routine Methods and Potential Benefits
J.Appl.Crystallogr., 47, 2014
|
|
4ZJ3
 
 | | Crystal structure of cephalexin bound acyl-enzyme intermediate of Val216AcrF mutant TEM1 beta-lactamase from Escherichia coli: E166N and V216AcrF mutant. | | Descriptor: | Beta-lactamase TEM | | Authors: | Xiao, H, Nasertorabi, F, Choi, S, Han, G.W, Reed, S.A, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploring the potential impact of an expanded genetic code on protein function.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3ZHH
 
 | | X-ray structure of the full-length beta-lactamase from M.tuberculosis | | Descriptor: | BETA-LACTAMASE, SULFATE ION | | Authors: | Feiler, C, Fisher, A.C, Marrichi, M.J, Wright, L, Schmidpeter, P.A.M, Blankenfeldt, W, Pavelka, M, DeLisa, M.P. | | Deposit date: | 2012-12-21 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Directed Evolution of Mycobacterium Tuberculosis Beta-Lactamase Reveals Gatekeeper Residue that Regulates Antibiotic Resistance and Catalytic Efficiency.
Plos One, 8, 2013
|
|
5A92
 
 | | 15K X-ray structure with Cefotaxime: Exploring the Mechanism of beta- Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97, CEFOTAXIME, C3' cleaved, ... | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
3M6H
 
 | | Crystal Structure of Post-isomerized Ertapenem Covalent Adduct with TB B-lactamase | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
3ZNY
 
 | | Crystal structure of the class A extended-spectrum beta-lactamase CTX- M-96, a natural D240G mutant derived from CTX-M-12 | | Descriptor: | CTX-M-12A ENZYME | | Authors: | Power, P, Herman, R, Bouillenne, F, Ghiglione, B, Rodriguez, M.M, Galleni, M, Gutkind, G, Charlier, P, Sauvage, E. | | Deposit date: | 2013-02-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Kinetic Insights Into the "Ceftazidimase" Behavior of the Extended-Spectrum Beta-Lactamase Ctx-M-96.
Biochemistry, 54, 2015
|
|
3MKF
 
 | | SHV-1 beta-lactamase complex with GB0301 | | Descriptor: | ({[(2R)-2-{[(4-ethyl-2,3-dioxo-3,4-dihydropyrazin-1(2H)-yl)carbonyl]amino}-2-(4-hydroxyphenyl)acetyl]amino}methyl)boronic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Ke, W. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3MKE
 
 | | SHV-1 beta-lactamase complex with LP06 | | Descriptor: | 2-[(~{Z})-[1-(2-azanyl-1,3-thiazol-4-yl)-2-oxidanylidene-2-[[(6~{S})-4,4,6-trimethyl-1,3,2-dioxaborinan-2-yl]methylamino]ethylidene]amino]oxy-2-methyl-propanoic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | van den Akker, F, Ke, W. | | Deposit date: | 2010-04-14 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3M6B
 
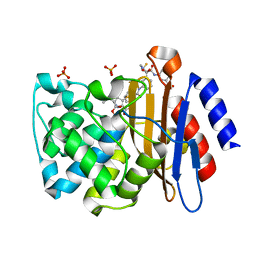 | | Crystal Structure of the Ertapenem Pre-isomerized Covalent Adduct with TB B-lactamase | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Fan, F, Blanchard, J.S. | | Deposit date: | 2010-03-15 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Biochemical and structural characterization of Mycobacterium tuberculosis beta-lactamase with the carbapenems ertapenem and doripenem.
Biochemistry, 49, 2010
|
|
5A90
 
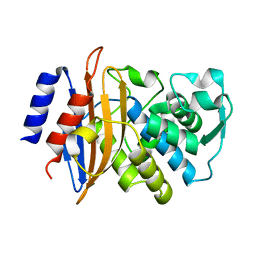 | | 100K Neutron Ligand Free: Exploring the Mechanism of beta-Lactam Ring Protonation in the Class A beta-lactamase Acylation Mechanism Using Neutron and X-ray Crystallography | | Descriptor: | BETA-LACTAMASE CTX-M-97 | | Authors: | Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Erskine, P.T, Tomanicek, S.J, Ostermann, A, Schrader, T.E, Ginell, S.L, Coates, L. | | Deposit date: | 2015-07-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (1.7 Å) | | Cite: | Exploring the Mechanism of Beta-Lactam Ring Protonation in the Class a Beta-Lactamase Acylation Mechanism Using Neutron and X-Ray Crystallography.
J.Med.Chem., 59, 2016
|
|
3MXR
 
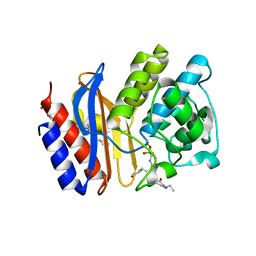 | | SHV-1 beta-lactamase complex with compound 1 | | Descriptor: | ({[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-4-(4-hydroxyphenyl)butanoyl]amino}methyl)boronic acid, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3N8S
 
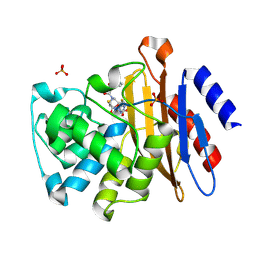 | | Crystal Structure of BlaC-E166A covalently bound with Cefamandole | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-hydroxy-2-phenylacetyl]amino}-2-oxoethyl]-5-{[(1-methyl-1H-tetrazol-5-yl)sulfanyl]methyl}-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Tremblay, L.W, Blanchard, J.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-11-24 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Michaelis Complex (1.2 A) and the Covalent Acyl Intermediate (2.0 A) of Cefamandole Bound in the Active Sites of the Mycobacterium tuberculosis beta-Lactamase K73A and E166A Mutants.
Biochemistry, 49, 2010
|
|
3MXS
 
 | | SHV-1 beta-lactamase complex with compound 2 | | Descriptor: | Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, N-[(dihydroxyboranyl)methyl]-Nalpha-[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]-D-tyrosinamide | | Authors: | Ke, W, van den Akker, F. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Novel Insights into the Mode of Inhibition of Class A SHV-1 {beta}-Lactamases Revealed by Boronic Acid Transition State Inhibitors.
Antimicrob.Agents Chemother., 55, 2011
|
|
3N6I
 
 | |
6WGR
 
 | | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase, GLYCEROL | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a beta-lactamase from Staphylococcus aureus subsp. aureus USA300_TCH1516
To Be Published
|
|
6WGP
 
 | | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of a beta lactamase from Xanthomonas campestris pv. campestris str. ATCC 33913
To Be Published
|
|
2OV5
 
 | | Crystal structure of the KPC-2 carbapenemase | | Descriptor: | BICINE, Carbapenemase | | Authors: | Ke, W, Bethel, C.R, Thomson, J.M, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of KPC-2: Insights into Carbapenemase Activity in Class A beta-Lactamases.
Biochemistry, 46, 2007
|
|
