1KRE
 
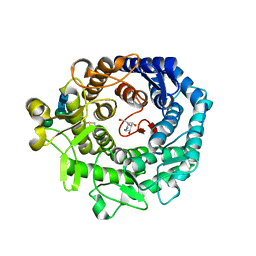 | | STRUCTURE OF P. CITRINUM ALPHA 1,2-MANNOSIDASE REVEALS THE BASIS FOR DIFFERENCES IN SPECIFICITY OF THE ER AND GOLGI CLASS I ENZYMES | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lobsanov, Y.D, Vallee, F, Imberty, A, Yoshida, T, Yip, P, Herscovics, A, Howell, P.L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes.
J.Biol.Chem., 277, 2002
|
|
3QEF
 
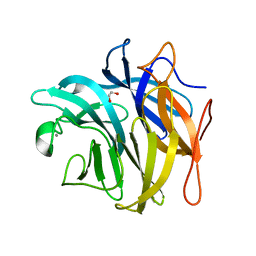 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3GUR
 
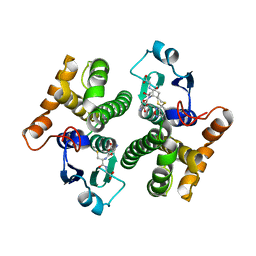 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
4JBX
 
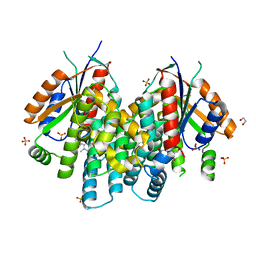 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with SK-78 | | Descriptor: | 1,2-ETHANEDIOL, 6-[3-hydroxy-2-(hydroxymethyl)prop-1-en-1-yl]-1,5-dimethylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, ... | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
2WM1
 
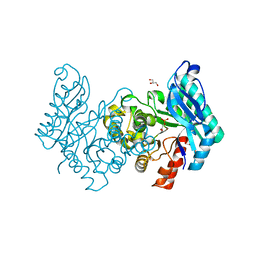 | | The crystal structure of human alpha-amino-beta-carboxymuconate- epsilon-semialdehyde decarboxylase in complex with 1,3- dihydroxyacetonephosphate suggests a regulatory link between NAD synthesis and glycolysis | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, 2-AMINO-3-CARBOXYMUCONATE-6-SEMIALDEHYDE DECARBOXYLASE, GLYCEROL, ... | | Authors: | Garavaglia, S, Perozzi, S, Galeazzi, L, Raffaelli, N, Rizzi, M. | | Deposit date: | 2009-06-29 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Crystal Structure of Human Alpha-Amino-Beta-Carboxymuconate-Epsilon-Semialdehyde Decarboxylase in Complex with 1,3-Dihydroxyacetonephosphate Suggests a Regulatory Link between Nad Synthesis and Glycolysis
FEBS J., 276, 2009
|
|
3L7D
 
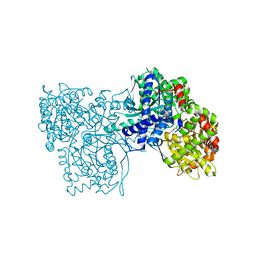 | | Crystal Structure of Glycogen Phosphorylase DK5 complex | | Descriptor: | 1-(2,3-dideoxy-3-fluoro-beta-D-arabino-hexopyranosyl)-4-[(phenylcarbonyl)amino]pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
3L7A
 
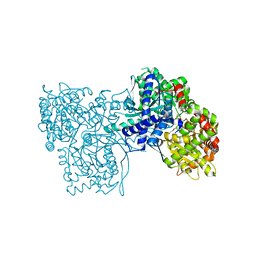 | | Crystal Structure of Glycogen Phosphorylase DK2 complex | | Descriptor: | 1-(3-deoxy-3-fluoro-beta-D-glucopyranosyl)-4-[(phenylcarbonyl)amino]pyrimidin-2(1H)-one, Glycogen phosphorylase, muscle form | | Authors: | Tsirkone, V.G, Lamprakis, C, Hayes, J.M, Skamnaki, V, Drakou, C, Zographos, S.E, Leonidas, D.D. | | Deposit date: | 2009-12-28 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1-(3-Deoxy-3-fluoro-beta-d-glucopyranosyl) pyrimidine derivatives as inhibitors of glycogen phosphorylase b: Kinetic, crystallographic and modelling studies.
Bioorg.Med.Chem., 18, 2010
|
|
1PPC
 
 | | GEOMETRY OF BINDING OF THE BENZAMIDINE-AND ARGININE-BASED INHIBITORS N-ALPHA-(2-NAPHTHYL-SULPHONYL-GLYCYL)-DL-P-AMIDINOPHENYLALANYL-PIPERIDINE (NAPAP) AND (2R,4R)-4-METHYL-1-[N-ALPHA-(3-METHYL-1,2,3,4-TETRAHYDRO-8-QUINOLINESULPHONYL)-L-ARGINYL]-2-PIPERIDINE CARBOXYLIC ACID (MQPA) TO HUMAN ALPHA-THROMBIN: X-RAY CRYSTALLOGRAPHIC DETERMINATION OF THE NAPAP-TRYPSIN COMPLEX AND MODELING OF NAPAP-THROMBIN AND MQPA-THROMBIN | | Descriptor: | 1-[N-(naphthalen-2-ylsulfonyl)glycyl-4-carbamimidoyl-D-phenylalanyl]piperidine, CALCIUM ION, TRYPSIN | | Authors: | Bode, W, Turk, D. | | Deposit date: | 1991-10-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Geometry of binding of the benzamidine- and arginine-based inhibitors N alpha-(2-naphthyl-sulphonyl-glycyl)-DL-p-amidinophenylalanyl-pipe ridine (NAPAP) and (2R,4R)-4-methyl-1-[N alpha-(3-methyl-1,2,3,4-tetrahydr quinolinesulphonyl)-L-arginyl]-2-piperidine carboxylic acid (MQPA) to human alpha-thrombin.X-ray crystallographic determination of the NAPAP-trypsin complex and modeling of NAPAP-thrombin and MQPA-thrombin.
Eur.J.Biochem., 193, 1990
|
|
4IG3
 
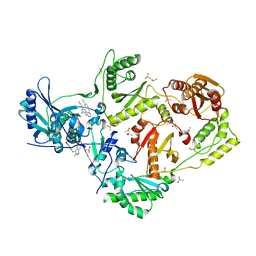 | | HIV-1 reverse transcriptase with bound fragment near Knuckles site | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
2WAJ
 
 | | Crystal structure of human Jnk3 complexed with a 1-aryl-3,4- dihydroisoquinoline inhibitor | | Descriptor: | 1-(3-BROMOPHENYL)-7-CHLORO-6-METHOXY-3,4-DIHYDROISOQUINOLINE, MITOGEN-ACTIVATED PROTEIN KINASE 10 | | Authors: | Bax, B.D, Christopher, J.A, Jones, E.J, Mosley, J.E. | | Deposit date: | 2009-02-08 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 1-Aryl-3,4-Dihydroisoquinoline Inhibitors of Jnk3.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1E48
 
 | |
1E47
 
 | |
3QD3
 
 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 1,1-Dimethylethyl {(3R,6S)-1-[2-amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-6-methyl-3-piperidinyl}carbamate | | Descriptor: | 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | Deposit date: | 2011-01-17 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
1FBH
 
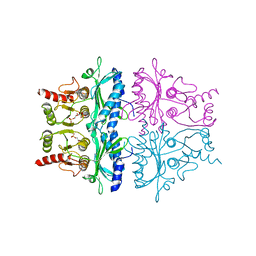 | | CRYSTALLOGRAPHIC STUDIES OF THE CATALYTIC MECHANISM OF THE NEUTRAL FORM OF FRUCTOSE-1,6-BISPHOSPHATASE | | Descriptor: | 1,6-di-O-phosphono-alpha-D-fructofuranose, 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Zhang, Y, Liang, J.-Y, Huang, S, Ke, H, Lipscomb, W.N. | | Deposit date: | 1992-10-16 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic studies of the catalytic mechanism of the neutral form of fructose-1,6-bisphosphatase.
Biochemistry, 32, 1993
|
|
4JPV
 
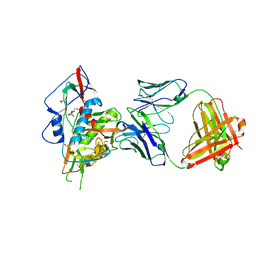 | | Crystal structure of broadly and potently neutralizing antibody 3bnc117 in complex with hiv-1 gp120 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2013-03-19 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.827 Å) | | Cite: | Somatic Mutations of the Immunoglobulin Framework Are Generally Required for Broad and Potent HIV-1 Neutralization.
Cell(Cambridge,Mass.), 153, 2013
|
|
7KRE
 
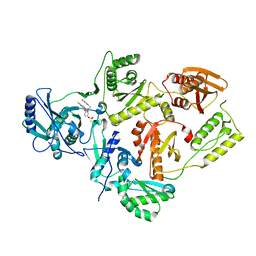 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-((6-cyanonaphthalen-1-yl)oxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ704) | | Descriptor: | 4-[(6-cyanonaphthalen-1-yl)oxy]-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.728 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
4KFB
 
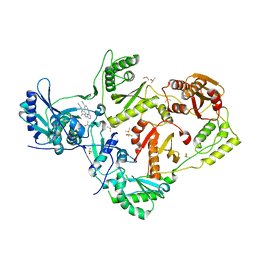 | | HIV-1 reverse transcriptase with bound fragment at NNRTI adjacent site | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
7KA1
 
 | | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 1.60 Angstrom resolution crystal structure of the beta-Q114A mutant Tryptophan Synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and cesium ion at the metal coordination site.
To be Published
|
|
7KRF
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-(3-(2-cyanovinyl)-5-fluorophenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ710) | | Descriptor: | 4-{3-[(E)-2-cyanoethenyl]-5-fluorophenoxy}-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
7KRC
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-4-(3-chloro-5-(2-cyanovinyl)phenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ709) | | Descriptor: | 4-{3-chloro-5-[(E)-2-cyanoethenyl]phenoxy}-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
7KRD
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-(3-chloro-5-cyanophenoxy)-3-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenyl sulfurofluoridate (JLJ702) | | Descriptor: | 4-(3-chloro-5-cyanophenoxy)-3-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenyl sulfurofluoridate, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Ippolito, J.A, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2020-11-19 | | Release date: | 2021-01-13 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Covalent Inhibition of Wild-Type HIV-1 Reverse Transcriptase Using a Fluorosulfate Warhead.
Acs Med.Chem.Lett., 12, 2021
|
|
4KB8
 
 | |
4I2Q
 
 | | Crystal structure of K103N/Y181C mutant of HIV-1 reverse transcriptase in complex with rilpivirine (TMC278) analogue | | Descriptor: | (2E)-3-(4-{[6-(1,3-benzothiazol-5-ylamino)-9H-purin-2-yl]amino}-3,5-dimethylphenyl)prop-2-enenitrile, 1,2-ETHANEDIOL, Gag-Pol polyprotein | | Authors: | Patel, D, Bauman, J.D, Das, K, Arnold, E. | | Deposit date: | 2012-11-22 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7004 Å) | | Cite: | A comparison of the ability of rilpivirine (TMC278) and selected analogues to inhibit clinically relevant HIV-1 reverse transcriptase mutants.
Retrovirology, 9, 2012
|
|
7JR6
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1-(3-fluorophenyl)-N-[trans-4-(2-hydroxypropan-2-yl)cyclohexyl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide, GLUTATHIONE, Hematopoietic prostaglandin D synthase, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
7JR8
 
 | | H-PDGS complexed with a 2-phenylimidazo[1,2-a]pyridine-6-carboxamide inhibitors | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLUTATHIONE, ... | | Authors: | Nolte, R.T, Somers, D.O, Gampe, R.T. | | Deposit date: | 2020-08-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | A knowledge-based, structural-aided discovery of a novel class of 2-phenylimidazo[1,2-a]pyridine-6-carboxamide H-PGDS inhibitors.
Bioorg.Med.Chem.Lett., 47, 2021
|
|
