8CKZ
 
 | |
8CKX
 
 | |
8CKV
 
 | |
8CL3
 
 | |
8CL0
 
 | |
8CKY
 
 | |
8CL2
 
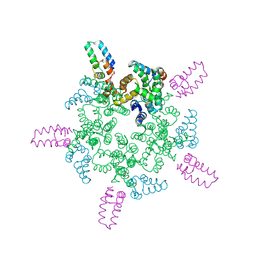 | |
8AW5
 
 | | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Heme O oxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hui, Z, Guoliang, Z. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of heme A synthase trimer from Aquifex aeolicus
To Be Published
|
|
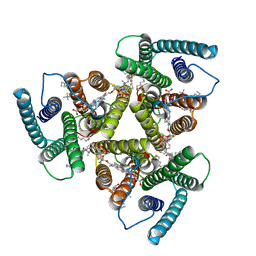
2CHM
 
 | | Crystal structure of N2 substituted pyrazolo pyrimidinones - a flipped binding mode in PDE5 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-[2-(BUT-3-EN-1-YLOXY)-5-(1-HYDROXYVINYL)PYRIDIN-3-YL]-3-ETHYL-2-(1-ETHYLAZETIDIN-3-YL)-1,2,6,7A-TETRAHYDRO-7H-PYRAZOLO[4,3-D]PYRIMIDIN-7-ONE, CGMP-SPECIFIC 3', ... | | Authors: | Allerton, C.M.N, Barber, C.G, Beaumont, K.C, Brown, D.G, Cole, S.M, Ellis, D, Lane, C.A.L, Maw, G.N, Mount, N.M, Rawson, D.J, Robinson, C.M, Street, S.D.A, Summerhill, N.W. | | Deposit date: | 2006-03-15 | | Release date: | 2006-06-08 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Novel Series of Potent and Selective Pde5 Inhibitors with Potential for High and Dose-Independent Oral Bioavailability
J.Med.Chem., 49, 2006
|
|
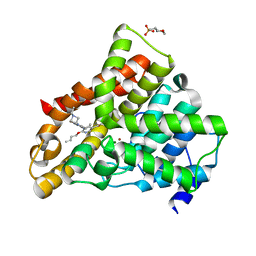
8BQ5
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 1 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1Z3N
 
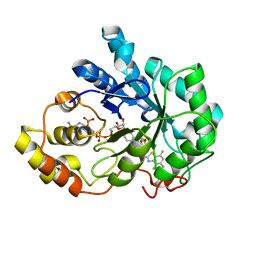 | | Human aldose reductase in complex with NADP+ and the inhibitor lidorestat at 1.04 angstrom | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase, {3-[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-1H-INDOL-1-YL}ACETIC ACID | | Authors: | Van Zandt, M.C, Jones, M.L, Gunn, D.E, Geraci, L.S, Jones, J.H, Sawicki, D.R, Sredy, J, Jacot, J.L, Dicioccio, A.T, Petrova, T, Mitschler, A, Podjarny, A.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Discovery of 3-[(4,5,7-trifluorobenzothiazol-2-yl)methyl]indole-N-acetic acid (lidorestat) and congeners as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications
J.Med.Chem., 48, 2005
|
|
8CIW
 
 | | Methylsuccinyl-CoA dehydrogenase from Pseudomonas migulae with bound FAD and (2S)-methylsuccinyl-CoA | | Descriptor: | (2S)-Methylsuccinyl-CoA, (2S)-methylsuccinyl-CoA dehydrogenase, CITRATE ANION, ... | | Authors: | Zarzycki, J, McLean, R, Erb, T.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Exploring alternative pathways for the in vitro establishment of the HOPAC cycle for synthetic CO 2 fixation.
Sci Adv, 9, 2023
|
|
8C6J
 
 | | Human spliceosomal PM5 C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Dybkov, O, Kastner, B, Luehrmann, R. | | Deposit date: | 2023-01-12 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Regulation of 3' splice site selection after step 1 of splicing by spliceosomal C* proteins.
Sci Adv, 9, 2023
|
|
8C07
 
 | |
8C06
 
 | |
8COI
 
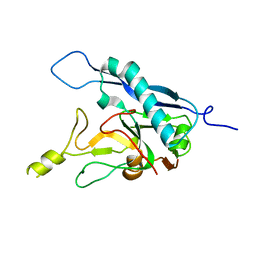 | | Human adenovirus-derived synthetic ADDobody binder | | Descriptor: | ADDobody | | Authors: | Buzas, D, Toelzer, C, Gupta, K, Berger-Schaffitzel, C, Berger, I. | | Deposit date: | 2023-02-28 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
8HQA
 
 | |
8HQ9
 
 | |
1Z28
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, Phenol-sulfating phenol sulfotransferase 1 | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1Z29
 
 | | Crystal Structures of SULT1A2 and SULT1A1*3: Implications in the bioactivation of N-hydroxy-2-acetylamino fluorine (OH-AAF) | | Descriptor: | ACETIC ACID, ADENOSINE-3'-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Lu, J, Li, H, Liu, M.C, Zhang, J, Li, M, An, X, Chang, W. | | Deposit date: | 2005-03-07 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of SULT1A2 and SULT1A1 *3: insights into the substrate inhibition and the role of Tyr149 in SULT1A2.
Biochem.Biophys.Res.Commun., 396, 2010
|
|
1YZI
 
 | |
2JS7
 
 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
2JQ9
 
 | | VPS4A MIT-CHMP1A complex | | Descriptor: | Chromatin-modifying protein 1a, Vacuolar protein sorting-associating protein 4A | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-05-30 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQH
 
 | | VPS4B MIT | | Descriptor: | Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-01 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
2JQK
 
 | | VPS4B MIT-CHMP2B Complex | | Descriptor: | Charged multivesicular body protein 2b, Vacuolar protein sorting-associating protein 4B | | Authors: | Stuchell-Brereton, M.D, Skalicky, J.J, Kieffer, C, Ghaffarian, S, Sundquist, W.I. | | Deposit date: | 2007-06-02 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | ESCRT-III recognition by VPS4 ATPases.
Nature, 449, 2007
|
|
