7AOC
 
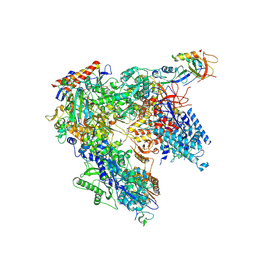 | | Schizosaccharomyces pombe RNA polymerase I (monomer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
7AOD
 
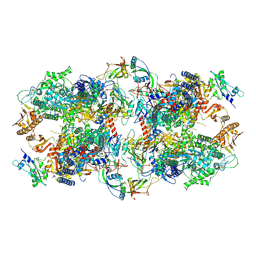 | | Schizosaccharomyces pombe RNA polymerase I (dimer) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit rpa1, DNA-directed RNA polymerase I subunit rpa14, ... | | Authors: | Heiss, F, Daiss, J, Becker, P, Engel, C. | | Deposit date: | 2020-10-14 | | Release date: | 2021-02-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Conserved strategies of RNA polymerase I hibernation and activation.
Nat Commun, 12, 2021
|
|
1BRG
 
 | |
8I4I
 
 | |
8I1Y
 
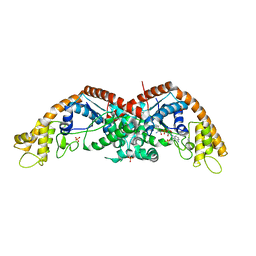 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I1W
 
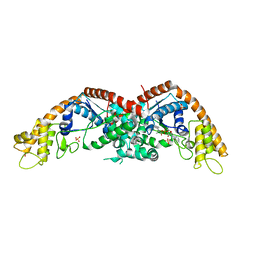 | |
6PK9
 
 | |
6N6C
 
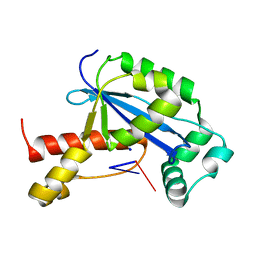 | | Vibrio cholerae Oligoribonuclease bound to pAA | | Descriptor: | RNA (5'-R(P*AP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.619 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
6N6E
 
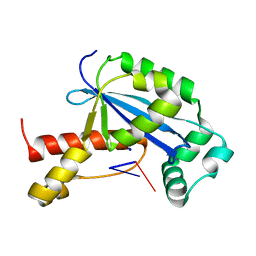 | | Vibrio cholerae Oligoribonuclease bound to pGA | | Descriptor: | RNA (5'-R(P*GP*A)-3'), RNA exonuclease 2 homolog,Small fragment nuclease, SODIUM ION | | Authors: | Lormand, J.D, Sondermann, H. | | Deposit date: | 2018-11-26 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.578 Å) | | Cite: | A dedicated diribonucleotidase resolves a key bottleneck for the terminal step of RNA degradation.
Elife, 8, 2019
|
|
1M0F
 
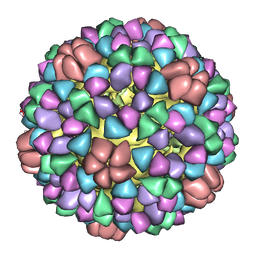 | | Structural Studies of Bacteriophage alpha3 Assembly, Cryo-electron microscopy | | Descriptor: | Capsid Protein F, Major Spike Protein G, Scaffolding Protein B, ... | | Authors: | Bernal, R.A, Hafenstein, S, Olson, N.H, Bowman, V.D, Chipman, P.R, Baker, T.S, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2002-06-12 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Structural Studies of Bacteriophage alpha3 Assembly
J.Mol.Biol., 325, 2003
|
|
5MWI
 
 | |
8BD6
 
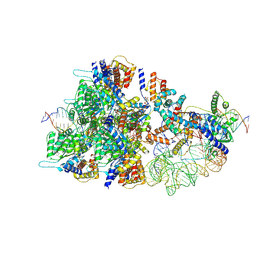 | | Cas12k-sgRNA-dsDNA-TnsC non-productive complex. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cas12k, DNA, ... | | Authors: | Schmitz, M, Querques, I, Oberli, S, Chanez, C, Jinek, M. | | Deposit date: | 2022-10-18 | | Release date: | 2022-12-28 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for the assembly of the type V CRISPR-associated transposon complex.
Cell, 185, 2022
|
|
8I9I
 
 | |
7V59
 
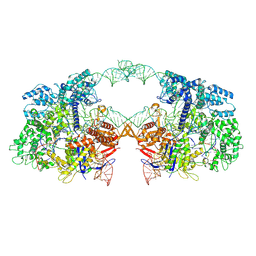 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | Authors: | Liu, J, Deng, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
2VRT
 
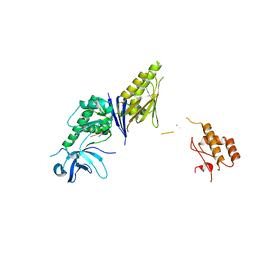 | | Crystal Structure of E. coli RNase E possessing M1 RNA fragments - Catalytic Domain | | Descriptor: | 5'-R(*UP*UP)-3', 5'-R(*UP*UP*GP)-3', RIBONUCLEASE E, ... | | Authors: | Koslover, D.J, Callaghan, A.J, Marcaida, M.J, Garman, E.F, Martick, M, Scott, W.G, Luisi, B.F. | | Deposit date: | 2008-04-14 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of the Escherichia Coli Rnase E Apoprotein and a Mechanism for RNA Degradation.
Structure, 16, 2008
|
|
7YYN
 
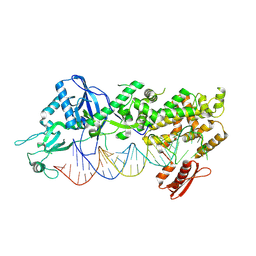 | | Mammalian Dicer in the dicing state with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Isoform 2 of Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
6EXZ
 
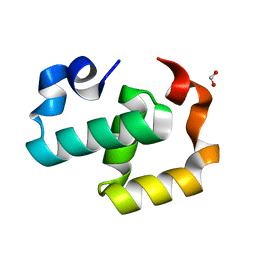 | | Crystal structure of Mex67 C-term | | Descriptor: | FORMIC ACID, mRNA export factor MEX67 | | Authors: | Mohamad, N, Bravo, J. | | Deposit date: | 2017-11-10 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mip6 binds directly to the Mex67 UBA domain to maintain low levels of Msn2/4 stress-dependent mRNAs.
Embo Rep., 2019
|
|
1C2P
 
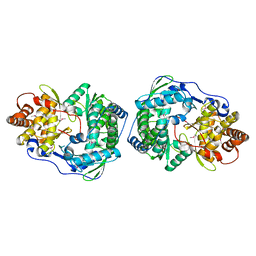 | | HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Lesburg, C.A, Cable, M.B, Ferrari, E, Hong, Z, Mannarino, A.F, Weber, P.C. | | Deposit date: | 1999-07-26 | | Release date: | 2000-04-05 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site.
Nat.Struct.Biol., 6, 1999
|
|
4YE6
 
 | |
4YE9
 
 | |
4YE8
 
 | |
1L9K
 
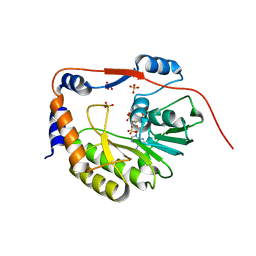 | | dengue methyltransferase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Egloff, M.P, Benarroch, D, Selisko, B, Romette, J.L, Canard, B. | | Deposit date: | 2002-03-25 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An RNA cap (nucleoside-2'-O-) methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization
Embo J., 21, 2002
|
|
8JHP
 
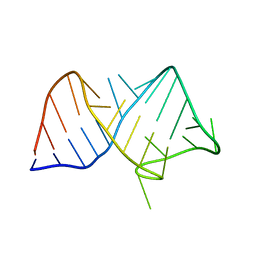 | |
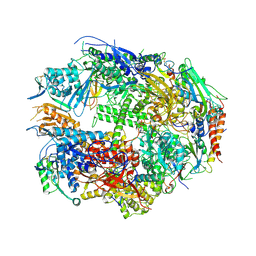
1TWG
 
 | | RNA polymerase II complexed with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
1TWH
 
 | | RNA polymerase II complexed with 2'dATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, ... | | Authors: | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
