2WKI
 
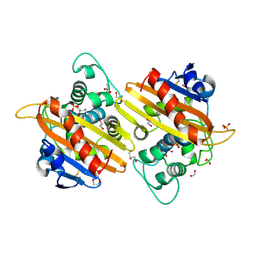 | | Crystal structure of the OXA-10 K70C mutant at pH 7.0 | | Descriptor: | 1,2-ETHANEDIOL, BETA-LACTAMASE OXA-10, GLYCEROL, ... | | Authors: | Vercheval, L, Bauvois, C, Kerff, F, Sauvage, E, Guiet, R, Charlier, P, Galleni, M. | | Deposit date: | 2009-06-11 | | Release date: | 2010-08-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three Factors that Modulate the Activity of Class D Beta-Lactamases and Interfere with the Post-Translational Carboxylation of Lys70.
Biochem.J., 432, 2010
|
|
3F1N
 
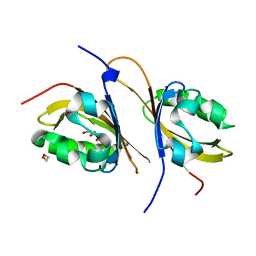 | | Crystal structure of a high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with internally bound ethylene glycol. | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7GYD
 
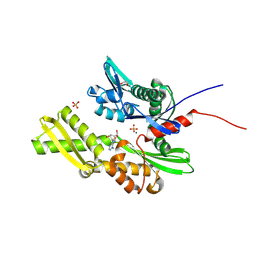 | | Crystal Structure of HSP72 in complex with ligand 10 at 11.40 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
7GYE
 
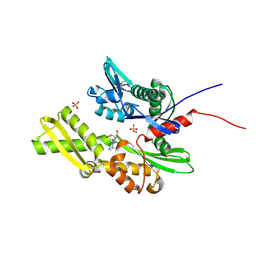 | | Crystal Structure of HSP72 in complex with ligand 10 at 12.54 MGy X-ray dose. | | Descriptor: | 1,2-ETHANEDIOL, 8-bromoadenosine, Heat shock 70 kDa protein 1A, ... | | Authors: | Cabry, M, Rodrigues, M.J, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2024-01-12 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Specific radiation damage to halogenated inhibitors and ligands in protein-ligand crystal structures.
J.Appl.Crystallogr., 57, 2024
|
|
4JP5
 
 | | X-ray structure of uridine phosphorylase from Yersinia pseudotuberculosis in unliganded state at 2.27 A resolution | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Balaev, V.V, Lashkov, A.A, Prokofev, I.I, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | X-ray structure of uridine phosphorylase from Yersinia pseudotuberculosis in unliganded state at 2.27 A resolution
To be Published
|
|
9MDD
 
 | | Crystal Structure of human cyclic GMP-AMP synthase (cGAS) in complex with compound 23; (S)-1-(6,7-dichloro-1-methyl-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl)-2-methoxyethan-1-one | | Descriptor: | 3-{[(2S)-1-{[(5P)-3-chloro-5-(1H-imidazol-2-yl)phenyl]amino}-1-oxopropan-2-yl](methyl)carbamoyl}pyridine-2-carboxylic acid, Cyclic GMP-AMP synthase, MANGANESE (II) ION, ... | | Authors: | Sietsema, D.V. | | Deposit date: | 2024-12-05 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyrimidine Amide cGAS Inhibitors via Structure-Guided Hybridization.
Acs Med.Chem.Lett., 15, 2024
|
|
1M4K
 
 | | Crystal structure of the human natural killer cell activator receptor KIR2DS2 (CD158j) | | Descriptor: | 1,2-ETHANEDIOL, KILLER CELL IMMUNOGLOBULIN-LIKE RECEPTOR 2DS2, SULFATE ION | | Authors: | Saulquin, X, Gastinel, L.N, Vivier, E. | | Deposit date: | 2002-07-03 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the human natural killer cell
activating receptor KIR2DS2 (CD158j)
J.EXP.MED., 197, 2003
|
|
4JRV
 
 | | Crystal structure of EGFR kinase domain in complex with compound 4c | | Descriptor: | 4-(dimethylamino)-N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]butanamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
4D0Z
 
 | | GalNAc-T2 crystal soaked with UDP-5SGalNAc, mEA2 and manganese (Higher resolution dataset) | | Descriptor: | (2R,3R,4R,5R,6R)-3-(acetylamino)-4,5-dihydroxy-6-(hydroxymethyl)tetrahydro-2H-thiopyran-2-yl [(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-5-thio-alpha-D-galactopyranose, ... | | Authors: | Lira-Navarrete, E, Iglesias-Fernandez, J, Zandberg, W.F, Companon, I, Kong, Y, Corzana, F, Pinto, B.M, Clausen, H, Peregrina, J.M, Vocadlo, D, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2014-04-30 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate-Guided Front-Face Reaction Revealed by Combined Structural Snapshots and Metadynamics for the Polypeptide N- Acetylgalactosaminyltransferase 2.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
9G4W
 
 | | Crystal structure of the human METTL3-METTL14 in complex with small molecule inhibitor Compound 59 | | Descriptor: | 4-[(3~{R})-3-(cyclopropylmethylamino)piperidin-1-yl]-1-[(1~{R})-1-[4-(6-pyrrolidin-1-ylpyrazin-2-yl)-1,2,3-triazol-1-yl]ethyl]pyridin-2-one, CALCIUM ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Dutheuil, G, Oukoloff, K. | | Deposit date: | 2024-07-16 | | Release date: | 2025-02-12 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery, Optimization, and Preclinical Pharmacology of EP652, a METTL3 Inhibitor with Efficacy in Liquid and Solid Tumor Models.
J.Med.Chem., 68, 2025
|
|
9EJI
 
 | | Peptide-independent T cell receptor recognition of HLA-DQ2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Lim, J.J, Loh, T.J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2024-11-27 | | Release date: | 2025-03-26 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A naturally selected alpha beta T cell receptor binds HLA-DQ2 molecules without co-contacting the presented peptide.
Nat Commun, 16, 2025
|
|
6F56
 
 | |
9G4S
 
 | | Crystal structure of the human METTL3-METTL14 in complex with small molecule inhibitor Compound 56 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-[(3~{R})-3-(cyclopropylmethylamino)piperidin-1-yl]-1-[(1~{R})-1-[4-[5-(dimethylamino)pyridin-3-yl]-1,2,3-triazol-1-yl]ethyl]pyridin-2-one, CALCIUM ION, ... | | Authors: | Dutheuil, G, Oukoloff, K. | | Deposit date: | 2024-07-16 | | Release date: | 2025-02-12 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Optimization, and Preclinical Pharmacology of EP652, a METTL3 Inhibitor with Efficacy in Liquid and Solid Tumor Models.
J.Med.Chem., 68, 2025
|
|
6LC9
 
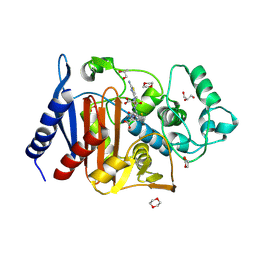 | | Crystal structure of AmpC Ent385 complex form with ceftazidime | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACYLATED CEFTAZIDIME, Beta-lactamase, ... | | Authors: | Kawai, A, Doi, Y. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis of Reduced Susceptibility to Ceftazidime-Avibactam and Cefiderocol inEnterobacter cloacaeDue to AmpC R2 Loop Deletion.
Antimicrob.Agents Chemother., 64, 2020
|
|
8DVL
 
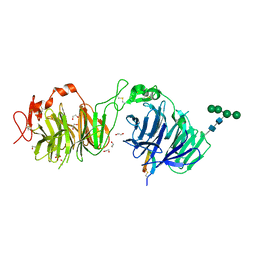 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.18 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
5CXX
 
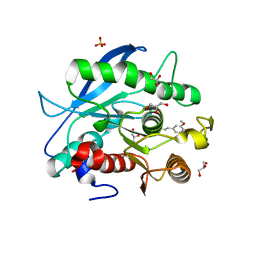 | | Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Gruninger, R.J, Abbott, D.W. | | Deposit date: | 2015-07-29 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Contributions of a unique beta-clamp to substrate recognition illuminates the molecular basis of exolysis in ferulic acid esterases.
Biochem.J., 473, 2016
|
|
8DVM
 
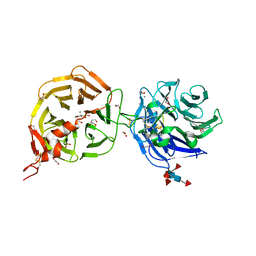 | | Crystal structure of LRP6 E3E4 in complex with disulfide constrained peptide E3.6 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Thakur, A.K, Liau, N.P.D, Sudhamsu, J, Hannoush, R.N. | | Deposit date: | 2022-07-29 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthetic Multivalent Disulfide-Constrained Peptide Agonists Potentiate Wnt1/ beta-Catenin Signaling via LRP6 Coreceptor Clustering.
Acs Chem.Biol., 18, 2023
|
|
2WNC
 
 | | Crystal structure of Aplysia ACHBP in complex with tropisetron | | Descriptor: | (3-ENDO)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL 1H-INDOLE-3-CARBOXYLATE, Soluble acetylcholine receptor | | Authors: | Sulzenbacher, G, Hibbs, R, Shi, J, Talley, T, Conrod, S, Kem, W, Taylor, P, Marchot, P, Bourne, Y. | | Deposit date: | 2009-07-08 | | Release date: | 2009-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor.
EMBO J., 28, 2009
|
|
4JWT
 
 | |
7YZ9
 
 | | Structure of catalytic domain of Rv1625c bound to nanobody NB4 | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, Adenylate cyclase, GLYCEROL, ... | | Authors: | Khanppnavar, B, Mehta, V.J, Iype, T, Korkhov, V.M. | | Deposit date: | 2022-02-19 | | Release date: | 2022-08-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of Mycobacterium tuberculosis Cya, an evolutionary ancestor of the mammalian membrane adenylyl cyclases.
Elife, 11, 2022
|
|
8K5O
 
 | | Cryo-EM structure of the RC-LH core comples from Halorhodospira halochloris | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-O-octyl-beta-D-glucopyranose, ... | | Authors: | Wang, G.-L, Qi, C.-H, Yu, L.-J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural insights into the unusual core photocomplex from a triply extremophilic purple bacterium, Halorhodospira halochloris.
J Integr Plant Biol, 66, 2024
|
|
4FNY
 
 | | Crystal structure of the R1275Q anaplastic lymphoma kinase catalytic domain in complex with a benzoxazole inhibitor | | Descriptor: | ALK tyrosine kinase receptor, N-(4-chlorophenyl)-5-[(6,7-dimethoxyquinolin-4-yl)oxy]-1,3-benzoxazol-2-amine | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The R1275Q Neuroblastoma Mutant and Certain ATP-competitive Inhibitors Stabilize Alternative Activation Loop Conformations of Anaplastic Lymphoma Kinase.
J.Biol.Chem., 287, 2012
|
|
7B47
 
 | | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Human p53DBD-R273H mutant bound to MQ: R273H-MQ (I) | | Descriptor: | (2~{R})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, (2~{S})-2-methyl-1-azabicyclo[2.2.2]octan-3-one, Cellular tumor antigen p53, ... | | Authors: | Degtjarik, O, Rozenberg, H, Shakked, Z. | | Deposit date: | 2020-12-02 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ).
Nat Commun, 12, 2021
|
|
2WQB
 
 | | Structure of the Tie2 kinase domain in complex with a thiazolopyrimidine inhibitor | | Descriptor: | 2-[3-(CYCLOHEXYLMETHYL)-5-PHENYL-IMIDAZOL-4-YL]-[1,3]THIAZOLO[4,5-E]PYRIMIDIN-7-AMINE, ANGIOPOIETIN-1 RECEPTOR | | Authors: | Brassington, C, Breed, J, Buttar, D, Fitzek, M, Forder, C, Hassall, L, Hayter, B.R, Jones, C.D, Luke, R.W.A, McCall, E, McCoull, W, Norman, R, Paterson, D, McMiken, H, Rowsell, S, Tucker, J.A. | | Deposit date: | 2009-08-18 | | Release date: | 2009-11-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Novel Thienopyrimidine and Thiazolopyrimidine Kinase Inhibitors with Activity Against Tie-2 in Vitro and in Vivo.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7ULB
 
 | | recombinant Mambalgin-1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Mambalgin-1 | | Authors: | Xu, J, Lei, X, Chen, L. | | Deposit date: | 2022-04-04 | | Release date: | 2023-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of recombinant mambalgin at 2.49 Angstroms resolution.
To Be Published
|
|
