7JQ4
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI7 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
6QXA
 
 | | Structure of membrane bound pyrophosphatase from Thermotoga maritima in complex with imidodiphosphate and N-[(2-amino-6-benzothiazolyl)methyl]-1H-indole-2-carboxamide (ATC) | | Descriptor: | (4S,5S)-1,2-DITHIANE-4,5-DIOL, IMIDODIPHOSPHORIC ACID, K(+)-stimulated pyrophosphate-energized sodium pump, ... | | Authors: | Vidilaseris, K, Goldman, A. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Asymmetry in catalysis byThermotoga maritimamembrane-bound pyrophosphatase demonstrated by a nonphosphorus allosteric inhibitor.
Sci Adv, 5, 2019
|
|
2VW0
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
4KTW
 
 | | Lactococcus phage 67 RuvC - apo form | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, RuvC endonuclease | | Authors: | Rafferty, J.B, Green, V. | | Deposit date: | 2013-05-21 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Mutants of phage bIL67 RuvC with enhanced Holliday junction binding selectivity and resolution symmetry.
Mol.Microbiol., 89, 2013
|
|
8JB3
 
 | |
6HA1
 
 | | Cryo-EM structure of a 70S Bacillus subtilis ribosome translating the ErmD leader peptide in complex with telithromycin | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3HXZ
 
 | | Crystal Structure of catalytic fragment of E. coli AlaRS G237A in complex with AlaSA | | Descriptor: | '5'-O-(N-(L-ALANYL)-SULFAMOYL)ADENOSINE, 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Guo, M, Yang, X.-L, Schimmel, P. | | Deposit date: | 2009-06-22 | | Release date: | 2009-12-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma.
Nature, 462, 2009
|
|
2VW2
 
 | | Crystal structure of the NanB sialidase from Streptococcus pneumoniae | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, SIALIDASE B | | Authors: | Xu, G, Potter, J.A, Russell, R.J.M, Oggioni, M.R, Andrew, P.W, Taylor, G.L. | | Deposit date: | 2008-06-13 | | Release date: | 2008-06-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Nanb Sialidase from Streptococcus Pneumoniae
J.Mol.Biol., 384, 2008
|
|
1GUD
 
 | | Hinge-bending motion of D-allose binding protein from Escherichia coli: three open conformations | | Descriptor: | D-ALLOSE-BINDING PERIPLASMIC PROTEIN, ZINC ION | | Authors: | Magnusson, U, Chaudhuri, B.N, Ko, J, Park, C, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2002-01-24 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of D-Allose Binding Protein from Escherichia Coli Bound to D-Allose at 1.8 A Resolution
J.Mol.Biol., 286, 1999
|
|
5DI9
 
 | |
1LN3
 
 | | Structure of Human Phosphatidylcholine Transfer Protein in Complex with Palmitoyl-Linoleoyl Phosphatidylcholine (Seleno-Met Protein) | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Phosphatidylcholine transfer protein | | Authors: | Roderick, S.L, Chan, W.W, Agate, D.S, Olsen, L.R, Vetting, M.W, Rajashankar, K.R, Cohen, D.E. | | Deposit date: | 2002-05-02 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of human phosphatidylcholine transfer protein in complex with its ligand.
Nat.Struct.Biol., 9, 2002
|
|
3I5N
 
 | | Crystal structure of c-Met with triazolopyridazine inhibitor 13 | | Descriptor: | 7-methoxy-N-[(6-phenyl[1,2,4]triazolo[4,3-b]pyridazin-3-yl)methyl]-1,5-naphthyridin-4-amine, Hepatocyte growth factor receptor | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.M, Boezio, A.A. | | Deposit date: | 2009-07-06 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and optimization of potent and selective triazolopyridazine series of c-Met inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5GMQ
 
 | | Structure of MERS-CoV RBD in complex with a fully human antibody MCA1 | | Descriptor: | 1,2-ETHANEDIOL, MCA1 heavy chain, MCA1 light chain, ... | | Authors: | Chen, C, Wang, J.M, Zou, T.T, Gao, X.P, Cui, S, Jin, Q. | | Deposit date: | 2016-07-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Human Neutralizing Monoclonal Antibody Inhibition of Middle East Respiratory Syndrome Coronavirus Replication in the Common Marmoset.
J. Infect. Dis., 215, 2017
|
|
5J1R
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one at 1.92A resolution | | Descriptor: | 3-(furan-3-yl)-1-(pyrrolidin-1-yl)propan-1-one, EthR | | Authors: | Blaszczyk, M, Surade, S, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2016-03-29 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Fragment-Sized EthR Inhibitors Exhibit Exceptionally Strong Ethionamide Boosting Effect in Whole-Cell Mycobacterium tuberculosis Assays.
ACS Chem. Biol., 12, 2017
|
|
4K1P
 
 | | Structure of the NheA component of the Nhe toxin from Bacillus cereus | | Descriptor: | 1,2-ETHANEDIOL, NheA, SULFATE ION | | Authors: | Ganash, M, Phung, D, Artymiuk, P.J. | | Deposit date: | 2013-04-05 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the NheA Component of the Nhe Toxin from Bacillus cereus: Implications for Function.
Plos One, 8, 2013
|
|
5DQ5
 
 | | Endothiapepsin in complex with fragment 209 | | Descriptor: | 3-[2-(4-methyl-1,2-oxazol-3-yl)ethyl]pyridine, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
to be published
|
|
2QT2
 
 | | Cyclized-Dehydrated Intermediate of GFP Variant Q183E in Chromophore Maturation | | Descriptor: | 1,2-ETHANEDIOL, Green fluorescent protein | | Authors: | Wood, T.I, Barondeau, D.P, Hitomi, C, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2007-07-31 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Kinetically Isolated Reaction Intermediates Provide Structural Characterization of the Green Fluorescence Protein Fluorophore Biosynthesis Pathway
To be Published
|
|
3HLA
 
 | | HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN A2.1 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2.1) (ALPHA CHAIN) | | Authors: | Saper, M.A, Bjorkman, P.J, Wiley, D.C. | | Deposit date: | 1989-10-06 | | Release date: | 1990-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution.
J.Mol.Biol., 219, 1991
|
|
5U2I
 
 | |
5DQF
 
 | | Horse Serum Albumin (ESA) in complex with Cetirizine | | Descriptor: | (2-{4-[(R)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, (2-{4-[(S)-(4-chlorophenyl)(phenyl)methyl]piperazin-1-yl}ethoxy)acetic acid, CHLORIDE ION, ... | | Authors: | Handing, K.B, Shabalin, I.G, Majorek, K.A, Chruszcz, M, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-14 | | Release date: | 2015-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of equine serum albumin in complex with cetirizine reveals a novel drug binding site.
Mol.Immunol., 71, 2016
|
|
6GNF
 
 | | Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, Glycogen synthase, ... | | Authors: | Cuesta-Seijo, J.A, Nielsen, M.M, Palcic, M.M. | | Deposit date: | 2018-05-30 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of theCatalyticDomain ofArabidopsis thalianaStarch Synthase IV, of Granule Bound Starch Synthase From CLg1 and of Granule Bound Starch Synthase I ofCyanophora paradoxaIllustrate Substrate Recognition in Starch Synthases.
Front Plant Sci, 9, 2018
|
|
3MPX
 
 | | Crystal structure of the DH and PH-1 domains of human FGD5 | | Descriptor: | FYVE, RhoGEF and PH domain-containing protein 5, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Nedyalkova, L, Tong, Y, Tempel, W, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the DH and PH-1 domains of human FGD5
TO BE PUBLISHED
|
|
7U1Y
 
 | | Structure of SPAC806.04c protein from fission yeast bound to AlF4 and Co2+ | | Descriptor: | COBALT (II) ION, Damage-control phosphatase SPAC806.04c, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Sanchez, A.M, Shuman, S. | | Deposit date: | 2022-02-22 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Fission yeast Duf89 and Duf8901 are cobalt/nickel-dependent phosphatase-pyrophosphatases that act via a covalent aspartyl-phosphate intermediate.
J.Biol.Chem., 298, 2022
|
|
5D3S
 
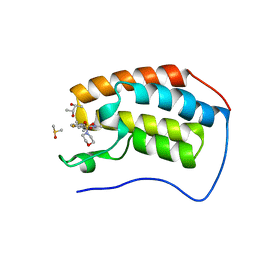 | | First bromodomain of BRD4 bound to inhibitor XD44 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-3-ethyl-N-[4-fluoro-3-(morpholin-4-ylsulfonyl)phenyl]-5-methyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M. | | Deposit date: | 2015-08-06 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
7TZZ
 
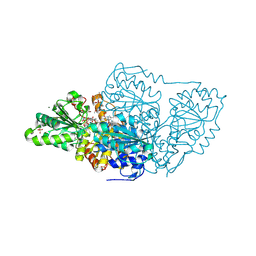 | | Crystal structure of arabidopsis thaliana acetohydroxyacid synthase P197T mutant in complex with bispyribac-sodium | | Descriptor: | 2,6-bis[(4,6-dimethoxypyrimidin-2-yl)oxy]benzoic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ... | | Authors: | Guddat, L.W, Cheng, Y. | | Deposit date: | 2022-02-16 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of resistance to herbicides that target acetohydroxyacid synthase.
Nat Commun, 13, 2022
|
|
