4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
2J3R
 
 | | The crystal structure of the bet3-trs31 heterodimer. | | Descriptor: | NITRATE ION, PALMITIC ACID, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 3, ... | | Authors: | Kim, Y.-G, Oh, B.-H. | | Deposit date: | 2006-08-23 | | Release date: | 2006-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Architecture of the Multisubunit Trapp I Complex Suggests a Model for Vesicle Tethering.
Cell(Cambridge,Mass.), 127, 2006
|
|
4ICC
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and JF0064 | | Descriptor: | 2,2',3,3',5,5',6,6'-octafluorobiphenyl-4,4'-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Porte, S, de Lera, A.R, Martin, M.J, de la Fuente, J.A, Klebe, G, Farres, J, Pares, X, Podjarny, A. | | Deposit date: | 2012-12-10 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Identification of a novel polyfluorinated compound as a lead to inhibit the human enzymes aldose reductase and AKR1B10: structure determination of both ternary complexes and implications for drug design.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6TRC
 
 | | Cryo- EM structure of the Thermosynechococcus elongatus photosystem I in the presence of cytochrome c6 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Koelsch, A, Radon, C, Baumert, A, Buerger, J, Mielke, T, Lisdat, F, Zouni, A, Wendler, P. | | Deposit date: | 2019-12-18 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Current limits of structural biology: The transient interaction between cytochrome c6 and photosystem I
Curr.Opin.Struct.Biol., 2, 2020
|
|
2YV5
 
 | | Crystal structure of Yjeq from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, YjeQ protein, ... | | Authors: | Wang, H, Kaminishi, T, Hanawa-Suetsugu, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of YjeQ from Aquifex aeolicus
To be Published
|
|
8WL2
 
 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CW state. | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-29 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WO5
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CCW state | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WLT
 
 | | Cryo-EM structure of the membrane-anchored part of the flagellar motor-hook complex in the CCW state | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar P-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-01 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WOE
 
 | | Cryo-EM structure of the intact flagellar motor-hook complex in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar L-ring protein, Flagellar M-ring protein, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-10-07 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
8WIW
 
 | | Cryo-EM structure of the flagellar C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2023-09-25 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
7RQ8
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7RQ9
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with iboxamycin, mRNA, deacylated A- and E-site tRNAs, and aminoacylated P-site tRNA at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Mitcheltree, M.J, Pisipati, A, Syroegin, E.A, Silvestre, K.J, Klepacki, D, Mason, J.D, Terwilliger, D.W, Testolin, G, Pote, A.R, Wu, K.J.Y, Ladley, R.P, Chatman, K, Mankin, A.S, Polikanov, Y.S, Myers, A.G. | | Deposit date: | 2021-08-06 | | Release date: | 2021-10-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A synthetic antibiotic class overcoming bacterial multidrug resistance.
Nature, 599, 2021
|
|
7RRO
 
 | | Structure of the 48-nm repeat doublet microtubule from bovine tracheal cilia | | Descriptor: | Armadillo repeat containing 4, Chromosome 3 C1orf194 homolog, Cilia and flagella associated protein 161, ... | | Authors: | Gui, M, Anderson, J.R, Botsch, J.J, Meleppattu, S, Singh, S.K, Zhang, Q, Brown, A. | | Deposit date: | 2021-08-10 | | Release date: | 2021-10-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | De novo identification of mammalian ciliary motility proteins using cryo-EM.
Cell, 184, 2021
|
|
7S3D
 
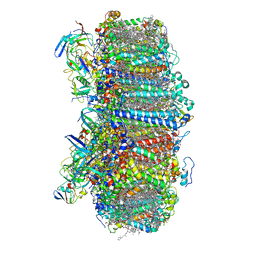 | | Structure of photosystem I with bound ferredoxin from Synechococcus sp. PCC 7335 acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2Fe-2S ferredoxin-type domain-containing protein, ... | | Authors: | Gisriel, C.J, Flesher, D.A, Shen, G, Wang, J, Ho, M, Brudvig, G.W, Bryant, D.A. | | Deposit date: | 2021-09-05 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a photosystem I-ferredoxin complex from a marine cyanobacterium provides insights into far-red light photoacclimation.
J.Biol.Chem., 298, 2021
|
|
7RQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MTI-tripeptidyl-tRNA analog ACCA-ITM, and chloramphenicol at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6POJ
 
 | |
6OLA
 
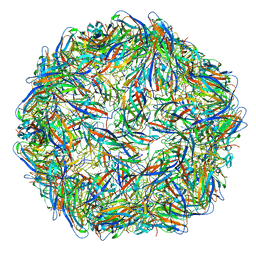 | | Structure of the PCV2d virus-like particle | | Descriptor: | Capsid protein, DNA (5'-D(P*CP*CP*GP*G)-3') | | Authors: | Khayat, R, Wen, K, Alimova, A, Galarza, J, Gottlieb, P. | | Deposit date: | 2019-04-16 | | Release date: | 2019-09-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural characterization of the PCV2d virus-like particle at 3.3 angstrom resolution reveals differences to PCV2a and PCV2b capsids, a tetranucleotide, and an N-terminus near the icosahedral 3-fold axes.
Virology, 537, 2019
|
|
4WZ7
 
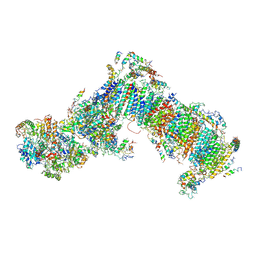 | | Crystal structure of mitochondrial NADH:ubiquinone oxidoreductase from Yarrowia lipolytica. | | Descriptor: | 39-kDa subunit, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Wirth, C, Zickermann, V, Brandt, U, Hunte, C. | | Deposit date: | 2014-11-18 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I.
Science, 347, 2015
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of the bacterial flagellar motor rotational switching.
Cell Res., 34, 2024
|
|
6ONY
 
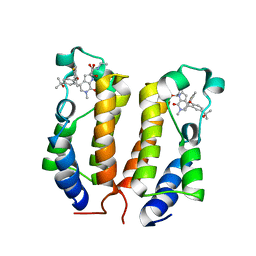 | | BRD2_Bromodomain1 complex with inhibitor 744 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, N-ethyl-4-[2-(4-fluoro-2,6-dimethylphenoxy)-5-(2-hydroxypropan-2-yl)phenyl]-6-methyl-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridine-2-carboxamide | | Authors: | Longenecker, K.L, Bigelow, L. | | Deposit date: | 2019-04-22 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer.
Nature, 578, 2020
|
|
8XJV
 
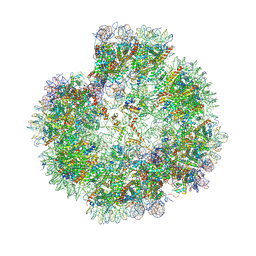 | | Structural basis for the linker histone H5-nucleosome binding and chromatin compaction | | Descriptor: | DNA, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, W.Y, Song, F, Zhu, P. | | Deposit date: | 2023-12-22 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for linker histone H5-nucleosome binding and chromatin fiber compaction.
Cell Res., 34, 2024
|
|
2YLO
 
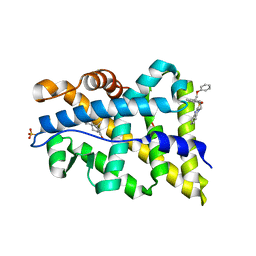 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 1-[2-(4-METHYLPHENOXY)ETHYL]-2-(2-PHENOXYETHYLSULFANYL)BENZIMIDAZOLE, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
2YLQ
 
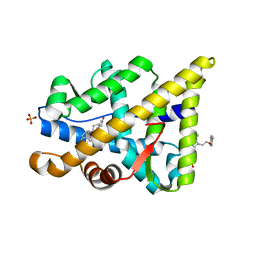 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[1-[2-(4-METHYLPHENOXY)ETHYL]BENZIMIDAZOL-2-YL]SULFANYLPROPANOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
2YLP
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[(2,4-DICHLOROPHENYL)METHYLSULFANYLMETHYL]BENZOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
