7B41
 
 | | Crystal structure of c-MET bound by compound 7 | | Descriptor: | 3-[(2-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7AK3
 
 | | CLK1 bound with CAF052 | | Descriptor: | Dual specificity protein kinase CLK1, ~{N}-[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]-4-pyrazolo[1,5-b]pyridazin-3-yl-pyrimidin-2-amine | | Authors: | Schroeder, M, Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-09-29 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Inhibitor Identifications Reveal Targeting Opportunity for the Atypical MAPK Kinase ERK3.
Int J Mol Sci, 21, 2020
|
|
6WBW
 
 | | Structure of Human HDAC2 in complex with an ethyl ketone inhibitor | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Klein, D.J, Yu, W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of ethyl ketone-based HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7AQ2
 
 | | Pseudomonas stutzeri nitrous oxide reductase mutant, H583A | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Bill, E, Kroneck, P.M.H, Einsle, O. | | Deposit date: | 2020-10-20 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Histidine-Gated Proton-Coupled Electron Transfer to the Cu A Site of Nitrous Oxide Reductase.
J.Am.Chem.Soc., 143, 2021
|
|
7AP1
 
 | | Klebsiella pneumoniae Seryl-tRNA synthetase in Complex with Compound SerS7HMDDA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine-tRNA ligase, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-10-15 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Synthesis and Biological Evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzimidazole Ribonucleoside Analogues as Aminoacyl-tRNA Synthetase Inhibitors.
Molecules, 25, 2020
|
|
6VT0
 
 | | Naegleria gruberi RNA ligase K170A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
6VTB
 
 | | Naegleria gruberi RNA ligase K326A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
7AWD
 
 | | Crystal structure of Peroxisome proliferator-activated receptor gamma (PPARG)in complex with garcinoic acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Endogenous vitamin E metabolites mediate allosteric PPAR gamma activation with unprecedented co-regulatory interactions.
Cell Chem Biol, 28, 2021
|
|
7AXF
 
 | |
6VW8
 
 | | Formate Dehydrogenase FdsABG subcomplex FdsBG from C. necator | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Young, T. | | Deposit date: | 2020-02-18 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and kinetic analyses of the FdsBG subcomplex of the cytosolic formate dehydrogenase FdsABG fromCupriavidus necator.
J.Biol.Chem., 295, 2020
|
|
7B06
 
 | | TgoT_RT521 apo | | Descriptor: | DNA polymerase | | Authors: | Samson, C, Legrand, P, Tekpinar, M, Rozenski, J, Abramov, M, Holliger, P, Pinheiro, V, Herdewijn, P, Delarue, M. | | Deposit date: | 2020-11-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Structural Studies of HNA Substrate Specificity in Mutants of an Archaeal DNA Polymerase Obtained by Directed Evolution.
Biomolecules, 10, 2020
|
|
6VPY
 
 | | I33M (I3.2 mutant from CH103 Lineage) | | Descriptor: | CHLORIDE ION, GLYCEROL, I33M heavy chain, ... | | Authors: | Fera, D, Zhou, J. | | Deposit date: | 2020-02-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The Effects of Framework Mutations at the Variable Domain Interface on Antibody Affinity Maturation in an HIV-1 Broadly Neutralizing Antibody Lineage.
Front Immunol, 11, 2020
|
|
7B1R
 
 | |
7B0T
 
 | | Crystal structure of MLLT1 YEATS domain T3 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, Protein ENL | | Authors: | Ni, X, Chaikuad, A, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-21 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
7AM3
 
 | | Crystal structure of Peptiligase mutant - M222P | | Descriptor: | GLYCEROL, SULFATE ION, Subtilisin BPN' | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7AM6
 
 | |
7AM8
 
 | | Crystal structure of Omniligase mutant W189F | | Descriptor: | ACRYLIC ACID, CHLORIDE ION, HISTIDINE, ... | | Authors: | Rozeboom, H.J, Janssen, D.J. | | Deposit date: | 2020-10-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | From thiol-subtilisin to omniligase: Design and structure of a broadly applicable peptide ligase.
Comput Struct Biotechnol J, 19, 2021
|
|
7ATI
 
 | | Crystal structure of dimeric chlorite dismutase variant Q74V (CCld Q74V) from Cyanothece sp. PCC7425 | | Descriptor: | Chlorite dismutase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schmidt, D, Mlynek, G, Djinovic-Carugo, K, Obinger, C. | | Deposit date: | 2020-10-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Arresting the Catalytic Arginine in Chlorite Dismutases: Impact on Heme Coordination, Thermal Stability, and Catalysis.
Biochemistry, 60, 2021
|
|
7B10
 
 | | Crystal structure of MLLT1 YEATS domain T1 mutant in complex with benzimidazole-amide based compound 1 | | Descriptor: | 1,2-ETHANEDIOL, 3-iodanyl-4-methyl-~{N}-[2-(piperidin-1-ylmethyl)-3~{H}-benzimidazol-5-yl]benzamide, IODIDE ION, ... | | Authors: | Chaikuad, A, Ni, X, Brennan, P.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-23 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure and Inhibitor Binding Characterization of Oncogenic MLLT1 Mutants.
Acs Chem.Biol., 16, 2021
|
|
7AM4
 
 | |
7AQF
 
 | |
7AM7
 
 | |
7B24
 
 | |
7AQG
 
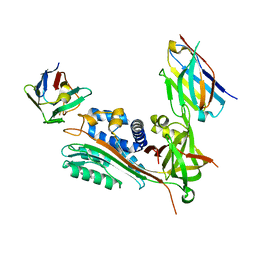 | | Crystal Structure of Small Molecule Inhibitor TM5484 Bound to Stabilized Active Plasminogen Activator Inhibitor-1 (PAI-1-W175F) | | Descriptor: | 5-Chloro-2-[[2-[3-(furan-3-yl)anilino]-2-oxoacetyl]amino]benzoic acid, Plasminogen activator inhibitor 1, VHH-2g-42 (Nb42), ... | | Authors: | Sillen, M, Strelkov, S.V, Declerck, P.J. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Insight into the Two-Step Mechanism of PAI-1 Inhibition by Small Molecule TM5484.
Int J Mol Sci, 22, 2021
|
|
7AVY
 
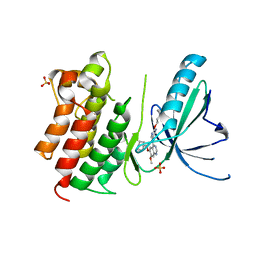 | | MerTK kinase domain in complex with quinazoline-based inhbitor | | Descriptor: | N-(2-(2-cyclopropylethoxy)pyrimidin-5-yl)-7-methoxy-6-(piperidin-4-ylmethoxy)quinazolin-4-amine, SULFATE ION, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
